
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,010,475 – 20,010,611 |
| Length | 136 |
| Max. P | 0.999875 |

| Location | 20,010,475 – 20,010,573 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20010475 98 + 22407834 GGGUAACUUCACUUGUGGUGGCUGCCGGUCUUCUUUAUUGUCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAA----------------------AGAGGAGCAGCUGCG .................(..(((((...((((((((.(((.....))).((((((..((((((.....)))))).))))))))----------------------)))))))))))..). ( -31.20) >DroSec_CAF1 188216 95 + 1 GGGACACUUCACUUGU---GGCUGCCUGUCUUCUUUAUUGUCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAA----------------------AGAGGAGCAGCUGCG ..............(.---.(((((...((((((((.(((.....))).((((((..((((((.....)))))).))))))))----------------------)))))))))))..). ( -31.20) >DroSim_CAF1 190732 95 + 1 GGAACACUUCACUUGU---GGCUGCCGGUCUUCUUUAUUGUCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAA----------------------AGAGGAGCAGCUGCG ..............(.---.(((((...((((((((.(((.....))).((((((..((((((.....)))))).))))))))----------------------)))))))))))..). ( -31.20) >DroEre_CAF1 185671 95 + 1 GGGACACUGCACUUGU---GGCUGCCGGUCUGCUUUAUUGUCACCCAACCAUUUGCAGCCAUUACUUAAAUGGCCCAAAUGAA----------------------AGAGGAGCAGCUGCG ..............(.---.(((((((((..((......)).)))....((((((..((((((.....)))))).))))))..----------------------....).)))))..). ( -27.70) >DroYak_CAF1 186623 95 + 1 UGGACAUUUCACUUGU---GGCUGCUGGCCUUCUUUAUUGUCACCCAACCAUUUGUAGCCAUUAUUUAAAUGGCCCAAAUGAA----------------------AGAAGAGCAGCUGCG ..............(.---.((((((...(((((((.(((.....))).((((((..((((((.....)))))).))))))))----------------------)))))))))))..). ( -31.20) >DroAna_CAF1 181127 117 + 1 GAGACUCUCCGCUUGU---UGCUGUCACUCUUCUUUAUUGUCGCUCAACAAUUUGCAGCCAUUAUUUAAAAGGCCCAGAUGGAGGAUGGCACUAUGGCAGUAGCCAGAGUAGCAGCUGGG ........(((.((((---((((((((..((((.....(((......)))(((((..(((...........))).)))))))))..))))....((((....)))).)))))))).))). ( -31.90) >consensus GGGACACUUCACUUGU___GGCUGCCGGUCUUCUUUAUUGUCACCCAACCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAA______________________AGAGGAGCAGCUGCG ...................((((((...((((((...(((.....))).((((((..((((((.....)))))).))))))........................))))))))))))... (-21.05 = -21.38 + 0.33)

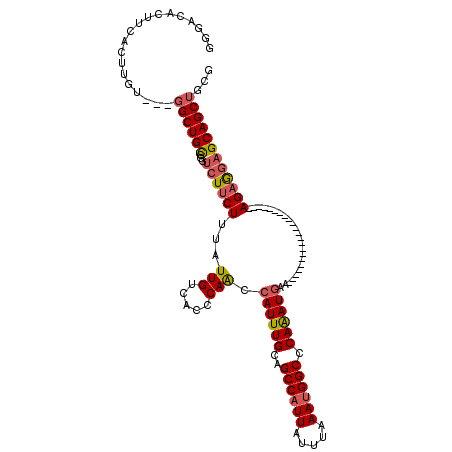

| Location | 20,010,475 – 20,010,573 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20010475 98 - 22407834 CGCAGCUGCUCCUCU----------------------UUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGACAAUAAAGAAGACCGGCAGCCACCACAAGUGAAGUUACCC ....((((((..(((----------------------(((((((((((((((.....))))))).))))))....(....)......)))))..))))))(((.....)))......... ( -30.50) >DroSec_CAF1 188216 95 - 1 CGCAGCUGCUCCUCU----------------------UUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGACAAUAAAGAAGACAGGCAGCC---ACAAGUGAAGUGUCCC (((.((((((..(((----------------------(((((((((((((((.....))))))).))))))....(....)......)))))..))))))(---(....))..))).... ( -31.10) >DroSim_CAF1 190732 95 - 1 CGCAGCUGCUCCUCU----------------------UUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGACAAUAAAGAAGACCGGCAGCC---ACAAGUGAAGUGUUCC (((.((((((..(((----------------------(((((((((((((((.....))))))).))))))....(....)......)))))..))))))(---(....))..))).... ( -30.60) >DroEre_CAF1 185671 95 - 1 CGCAGCUGCUCCUCU----------------------UUCAUUUGGGCCAUUUAAGUAAUGGCUGCAAAUGGUUGGGUGACAAUAAAGCAGACCGGCAGCC---ACAAGUGCAGUGUCCC .(((((((((..(((----------------------..(((((((((((((.....))))))).))))))(((.(....).....))))))..)))))).---.....)))........ ( -29.40) >DroYak_CAF1 186623 95 - 1 CGCAGCUGCUCUUCU----------------------UUCAUUUGGGCCAUUUAAAUAAUGGCUACAAAUGGUUGGGUGACAAUAAAGAAGGCCAGCAGCC---ACAAGUGAAAUGUCCA .((.((((..(((((----------------------(((((((((((((((.....))))))).))))))((((.....)))))))))))..)))).)).---................ ( -32.10) >DroAna_CAF1 181127 117 - 1 CCCAGCUGCUACUCUGGCUACUGCCAUAGUGCCAUCCUCCAUCUGGGCCUUUUAAAUAAUGGCUGCAAAUUGUUGAGCGACAAUAAAGAAGAGUGACAGCA---ACAAGCGGAGAGUCUC .....((((((((.((((....)))).)))((..(((((..(((.((((...........))))....((((((....))))))..))).))).))..)).---...)))))........ ( -30.20) >consensus CGCAGCUGCUCCUCU______________________UUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGACAAUAAAGAAGACCGGCAGCC___ACAAGUGAAGUGUCCC ....((((((..(((.......................((((((((((((((.....))))))).)))))))...(....)........)))..)))))).................... (-23.76 = -24.18 + 0.42)

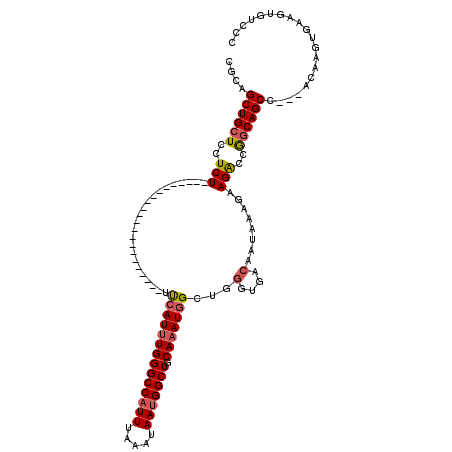

| Location | 20,010,515 – 20,010,611 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -27.70 |
| Energy contribution | -28.66 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20010515 96 + 22407834 UCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAAAGAGGAGCAGCUGCGUCUGCGAAAGCCUCCGUUUUGGGCGACGGGCUGUGGAU ...(((((((..((((.((((((.....))))))(((((.....((((.((((......)))).....))))...)))))))))..))))).)).. ( -37.70) >DroSec_CAF1 188253 96 + 1 UCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAAAGAGGAGCAGCUGCGUCUGCGGAAGCCUCCGUUUUGGCUGACUGGUUGUGCAU .......((((((((..((((((.....)))))).))))))........(((((((.(((.((.(((((....))))).)).)))))))))))).. ( -31.00) >DroSim_CAF1 190769 96 + 1 UCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAAAGAGGAGCAGCUGCGUCUGCGGAAGCCUCCGUUUUGGCUGACUGGUUGUGGAU ...((((((((...(((((((((.....))))((((..........)).)).)))))(((.((.(((((....))))).)).))))))))).)).. ( -33.90) >DroEre_CAF1 185708 96 + 1 UCACCCAACCAUUUGCAGCCAUUACUUAAAUGGCCCAAAUGAAAGAGGAGCAGCUGCGUCUGCGGAAGCCUCCGUUUUGGGUGACUGGUGGUGGAU ((((((((.((((((..((((((.....)))))).)))))).....(((((..((((....))))..).))))...))))))))............ ( -35.20) >DroYak_CAF1 186660 96 + 1 UCACCCAACCAUUUGUAGCCAUUAUUUAAAUGGCCCAAAUGAAAGAAGAGCAGCUGCGUCUGCGGAAGCCUCCGUUUUCGGUGACAGGUUGUGGAU ...((((((((((((..((((((.....)))))).)))))(((((..((((..((((....))))..).)))..)))))(....).))))).)).. ( -32.20) >consensus UCACCCAGCCAUUUGCAGCCAUUAUUUAAAUGGCCCAAAUGAAAGAGGAGCAGCUGCGUCUGCGGAAGCCUCCGUUUUGGGUGACUGGUUGUGGAU ((((((((.((((((..((((((.....)))))).)))))).....(((((..((((....))))..).))))...))))))))............ (-27.70 = -28.66 + 0.96)

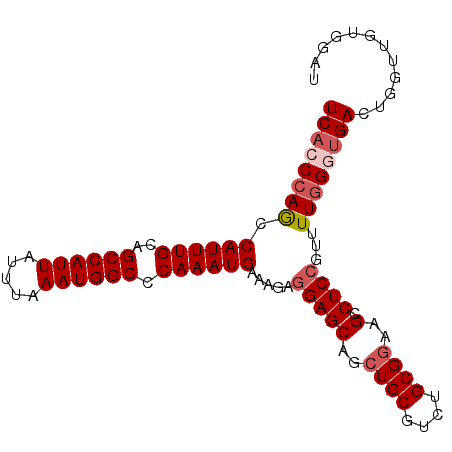
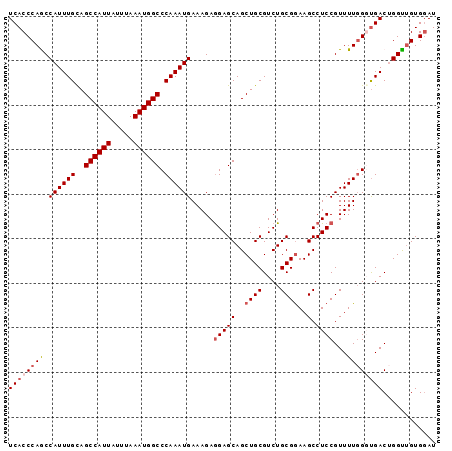
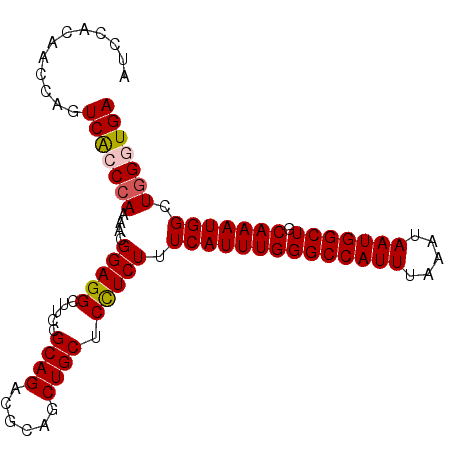
| Location | 20,010,515 – 20,010,611 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20010515 96 - 22407834 AUCCACAGCCCGUCGCCCAAAACGGAGGCUUUCGCAGACGCAGCUGCUCCUCUUUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGA ............(((((((....(((((.....((((......)))).))))).((((((((((((((.....))))))).))))))).))))))) ( -34.40) >DroSec_CAF1 188253 96 - 1 AUGCACAACCAGUCAGCCAAAACGGAGGCUUCCGCAGACGCAGCUGCUCCUCUUUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGA .........((.((((((.....(((((.....((((......)))).)))))...((((((((((((.....))))))).))))))))))).)). ( -34.00) >DroSim_CAF1 190769 96 - 1 AUCCACAACCAGUCAGCCAAAACGGAGGCUUCCGCAGACGCAGCUGCUCCUCUUUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGA .........((.((((((.....(((((.....((((......)))).)))))...((((((((((((.....))))))).))))))))))).)). ( -34.00) >DroEre_CAF1 185708 96 - 1 AUCCACCACCAGUCACCCAAAACGGAGGCUUCCGCAGACGCAGCUGCUCCUCUUUCAUUUGGGCCAUUUAAGUAAUGGCUGCAAAUGGUUGGGUGA ............((((((((...(((((.....((((......)))).))))).((((((((((((((.....))))))).))))))))))))))) ( -35.90) >DroYak_CAF1 186660 96 - 1 AUCCACAACCUGUCACCGAAAACGGAGGCUUCCGCAGACGCAGCUGCUCUUCUUUCAUUUGGGCCAUUUAAAUAAUGGCUACAAAUGGUUGGGUGA ..((.(((((.(((.(((....))).)))....((((......)))).........((((((((((((.....))))))).))))))))))))... ( -30.50) >consensus AUCCACAACCAGUCACCCAAAACGGAGGCUUCCGCAGACGCAGCUGCUCCUCUUUCAUUUGGGCCAUUUAAAUAAUGGCUGCAAAUGGCUGGGUGA ............(((((((....(((((.....((((......)))).))))).((((((((((((((.....))))))).))))))).))))))) (-30.70 = -30.98 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:32 2006