
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 20,007,395 – 20,007,533 |
| Length | 138 |
| Max. P | 0.991183 |

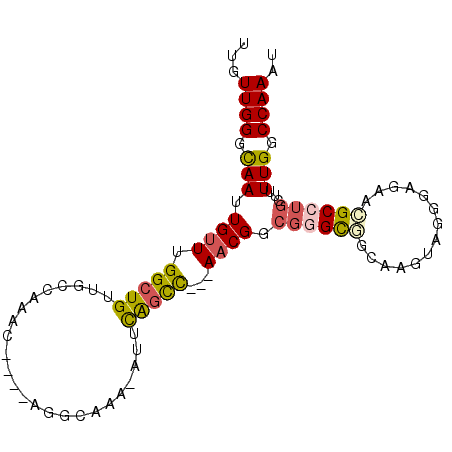
| Location | 20,007,395 – 20,007,493 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20007395 98 + 22407834 UUGUUGGGCAAUUGUUUGGCUGUUGCCAAACAUCCAGGCAAACAUUCAGCCAACAACGUCGGGCGGCGAGUUGGGAGAACGCCUGCUUUUGCCCAAAU ...((((((((.((((((((....))))))))..(((((.....(((..(((((..((((....)))).)))))..))).)))))...)))))))).. ( -41.80) >DroSec_CAF1 185149 83 + 1 UUGUUGGGCAAUUGGUUGGCUGUUGCCAAAC------------AUUCAGCC---AACGUCGGGCGGCGAGUUGGGAGAACGCCUGCUUUUGGCCAAAU ...((((.(((..(((((((((.((.....)------------)..)))))---)))..((((((..............)))))))..))).)))).. ( -30.74) >DroSim_CAF1 187655 83 + 1 UUGUUGGGCAAUUGUUUGGCUGUUGCCAAAC------------AUUCAGCC---AACGUCGGGCGGCGAGUUGGGAGAACGCCUGCUUUUGGCCAAAU ..(((((.....((((((((....)))))))------------).....))---)))....(((.(.((((.((.......)).)))).).))).... ( -30.30) >DroEre_CAF1 182585 94 + 1 UUGUUGGGCAAAUGUUUGGCUGUUGCCAAACAUCCAGGCAAACAUUCAGCC---AACGGCG-GCCGCAAGUACGGAGAACGCCUGCUUUUGGCCAAAU ((((..((...(((((((((....)))))))))))..)))).......(((---...)))(-((((.(((((.((......))))))).))))).... ( -36.40) >DroYak_CAF1 183543 95 + 1 UUGUUGGGCAAUUGUUUGGCUGUUGCCAACCAUCCAGGCAAAAAUGCAGUC---AACGGCGGGCCCCAAGUACGGAGAUCGCCUGUUUUUGGCCAAAU ...((((.(((....((((((((((((.........)))).....))))))---)).((((..(.((......)).)..)))).....))).)))).. ( -31.00) >DroAna_CAF1 179487 84 + 1 UUGUUGGGUAAUUGUUUGGCUGUC-----------UGGCUGUUUGUUGCCU---AACGGAAAGUAGCAAGCAUCGAGAAUGCCAACUUUUGGCCAAAU ...((((.(((..(((.(((..((-----------(.(.((((((((((..---........)))))))))).).)))..))))))..))).)))).. ( -29.60) >consensus UUGUUGGGCAAUUGUUUGGCUGUUGCCAAAC____AGGCAAA_AUUCAGCC___AACGGCGGGCGGCAAGUAGGGAGAACGCCUGCUUUUGGCCAAAU ...((((.(((.((((.(((((........................)))))...)))).((((((..............))))))...))).)))).. (-15.45 = -15.78 + 0.34)


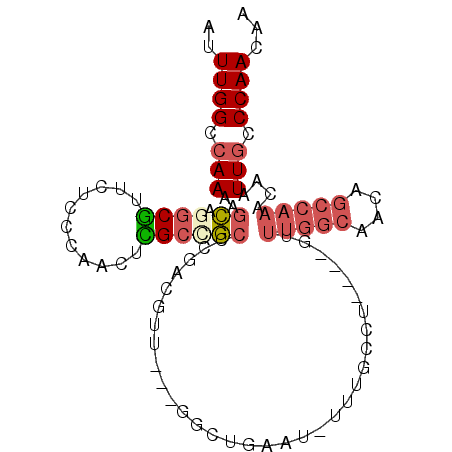
| Location | 20,007,395 – 20,007,493 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.93 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.31 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20007395 98 - 22407834 AUUUGGGCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUUGUUGGCUGAAUGUUUGCCUGGAUGUUUGGCAACAGCCAAACAAUUGCCCAACAA ..((((((((..((((((((((..(((((.((.(....).))..)))))..)))))))))).....((((((((....)))))))).))))))))... ( -45.20) >DroSec_CAF1 185149 83 - 1 AUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU---GGCUGAAU------------GUUUGGCAACAGCCAACCAAUUGCCCAACAA ..((((.(((....((((((((..((((((((.....))).)))---))..))))------------))))(((....)))......))).))))... ( -26.00) >DroSim_CAF1 187655 83 - 1 AUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU---GGCUGAAU------------GUUUGGCAACAGCCAAACAAUUGCCCAACAA .((..(((((...(.((((..............)))).)...))---)))..))(------------(((((((....))))))))............ ( -28.24) >DroEre_CAF1 182585 94 - 1 AUUUGGCCAAAAGCAGGCGUUCUCCGUACUUGCGGC-CGCCGUU---GGCUGAAUGUUUGCCUGGAUGUUUGGCAACAGCCAAACAUUUGCCCAACAA .((..(((((.....((((....((((....)))).-)))).))---)))..))((((.((..(((((((((((....)))))))))))))..)))). ( -37.50) >DroYak_CAF1 183543 95 - 1 AUUUGGCCAAAAACAGGCGAUCUCCGUACUUGGGGCCCGCCGUU---GACUGCAUUUUUGCCUGGAUGGUUGGCAACAGCCAAACAAUUGCCCAACAA ..((((.(((.(((.((((..(((((....)))))..)))))))---....(((....))).((..((((((....))))))..)).))).))))... ( -32.90) >DroAna_CAF1 179487 84 - 1 AUUUGGCCAAAAGUUGGCAUUCUCGAUGCUUGCUACUUUCCGUU---AGGCAACAAACAGCCA-----------GACAGCCAAACAAUUACCCAACAA .((((((....(((.((((((...)))))).))).......(((---.(((........))).-----------))).)))))).............. ( -21.50) >consensus AUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU___GGCUGAAU_UUUGCCU____GUUUGGCAACAGCCAAACAAUUGCCCAACAA ..((((.(((..((.((((...........)))))).................................(((((....)))))....))).))))... (-10.02 = -10.93 + 0.92)


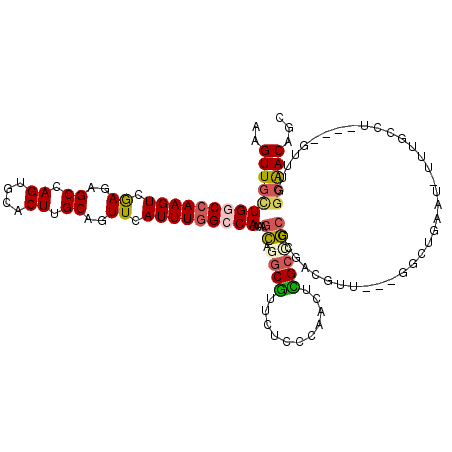
| Location | 20,007,413 – 20,007,533 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -20.16 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 20007413 120 - 22407834 AAGUUGCUGGCCAAGUCGAGUGCCAGUGCACUUGCAGUUCAUUUGGGCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUUGUUGGCUGAAUGUUUGCCUGGAUGUUUGGCAACAGC .....((((((((((.(((((((....))))))))....((((..(((((......((((((..(((((.((.(....).))..)))))..)))))))))))..)))).)))))..)))) ( -46.60) >DroSec_CAF1 185167 105 - 1 AAGUUGCUGGCCAAGUCGAGAGCCAGUGCACUUGCAGUUCAUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU---GGCUGAAU------------GUUUGGCAACAGC ..((((((((((((((...((((...(((....))))))))))))))))....(((((((((..((((((((.....))).)))---))..))))------------))))))))))... ( -44.10) >DroSim_CAF1 187673 105 - 1 AAGUUGCUGGCCAAGUCGAGAGCCAGUGCACUUGCAGUUCAUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU---GGCUGAAU------------GUUUGGCAACAGC ..((((((((((((((...((((...(((....))))))))))))))))....(((((((((..((((((((.....))).)))---))..))))------------))))))))))... ( -44.10) >DroEre_CAF1 182603 116 - 1 AAGUUGUUGGCCAAGUCGAGAGCCAGUGCACUUGCAGUUCAUUUGGCCAAAAGCAGGCGUUCUCCGUACUUGCGGC-CGCCGUU---GGCUGAAUGUUUGCCUGGAUGUUUGGCAACAGC ..(((((((.(((((.((....(((((((....)))((.((((..(((((.....((((....((((....)))).-)))).))---)))..))))...)))))).)))))))))))))) ( -45.50) >DroYak_CAF1 183561 117 - 1 AAGUUGUUGGCCAAGUCGAGAGCCAGUGCACUUGCAGUUCAUUUGGCCAAAAACAGGCGAUCUCCGUACUUGGGGCCCGCCGUU---GACUGCAUUUUUGCCUGGAUGGUUGGCAACAGC ..(((((..((((...(....)((((.(((..(((((((....((........))((((..(((((....)))))..))))...---)))))))....))))))).))))..)))))... ( -47.20) >DroAna_CAF1 179505 104 - 1 AAGUUUCUGACAAAGUUAAGAGCCAG--CACUUGCAGUUCAUUUGGCCAAAAGUUGGCAUUCUCGAUGCUUGCUACUUUCCGUU---AGGCAACAAACAGCCA-----------GACAGC ......(((..........((((..(--(....)).)))).((((((....(((.((((((...)))))).))).......(((---.(....).))).))))-----------))))). ( -24.50) >consensus AAGUUGCUGGCCAAGUCGAGAGCCAGUGCACUUGCAGUUCAUUUGGCCAAAAGCAGGCGUUCUCCCAACUCGCCGCCCGACGUU___GGCUGAAU_UUUGCCU____GUUUGGCAACAGC ..((((((((((((((.((..((.((....)).))..)).)))))))))...((.((((...........))))))....................................)))))... (-20.16 = -20.47 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:27 2006