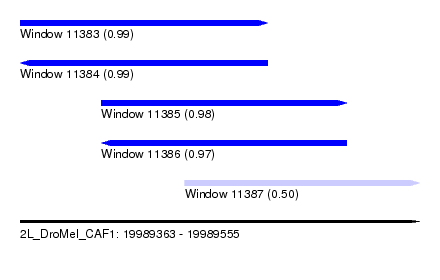
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,989,363 – 19,989,555 |
| Length | 192 |
| Max. P | 0.994264 |

| Location | 19,989,363 – 19,989,482 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -36.70 |
| Energy contribution | -36.22 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19989363 119 + 22407834 GAAGUGAGUACUAAAUGCACCCAGCUGGCGCUCAUAUU-UGUAUGUUCGCUAAUUUGCAGCUGAAUGGAGCUUAAACGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((.....(((.(.(((((((((..((((..-..))))..))))..((((((((((..(.........)..))))))))))......))))).)))).......)))).)))) ( -35.90) >DroSec_CAF1 172148 119 + 1 GAAGUGAGUACUAAAUGCACCCAGCUGGCGCUCAUAUU-UGUAUGUUCGCUAAUUUGCAGCUGAAUGUCGCUUAAAUGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((.....(((.(.(((((((((..((((..-..))))..))))..((((((((((.((.........)).))))))))))......))))).)))).......)))).)))) ( -36.40) >DroSim_CAF1 174718 119 + 1 GAAGUGAGUACUAAAUGCACCCAGCUGGCGCUCAUAUU-UGUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAAUGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((.....(((.(.(((((((((..((((..-..))))..))))..((((((((((.((.........)).))))))))))......))))).)))).......)))).)))) ( -36.40) >DroEre_CAF1 170215 120 + 1 GAAGUGAGUACUAAAUGCGCCCAGCUGGCGUUCAUACUCCGUGUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAGACGCAGCUGCGGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((....((..((((((((((((..(((((...)))))..))))..((((((((((.....(.....)...))))))))))......))))).)))..))....)))).)))) ( -40.70) >DroYak_CAF1 171042 120 + 1 GAAGUGAGUACUAAAUGCGCCCAGCUGGCGCUCAUAUUUUGUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAGCGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((....((..((((((((((((..(((((...)))))..))))..(((((((((.....((((....)))))))))))))......))))).)))..))....)))).)))) ( -42.60) >consensus GAAGUGAGUACUAAAUGCACCCAGCUGGCGCUCAUAUU_UGUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAACGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUU ..((((((((.....(((.(.(((((((((..(((((...)))))..))))..((((((((((...............))))))))))......))))).)))).......)))).)))) (-36.70 = -36.22 + -0.48)

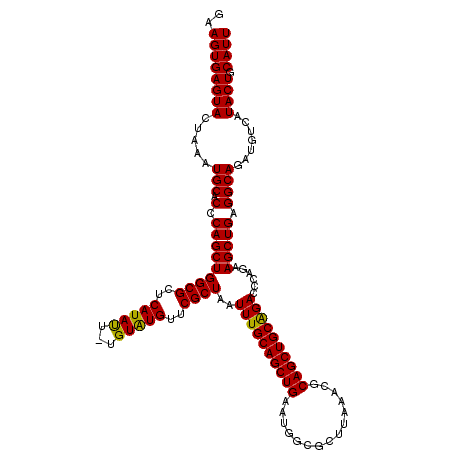
| Location | 19,989,363 – 19,989,482 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -33.92 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19989363 119 - 22407834 AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGUUUAAGCUCCAUUCAGCUGCAAAUUAGCGAACAUACA-AAUAUGAGCGCCAGCUGGGUGCAUUUAGUACUCACUUC ..((.(((((......((((((((((....(((.((((((((.((.........)).)))))))).))).(((..((((..-..))))..))).))))))).)))....))))))).... ( -35.60) >DroSec_CAF1 172148 119 - 1 AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCAUUUAAGCGACAUUCAGCUGCAAAUUAGCGAACAUACA-AAUAUGAGCGCCAGCUGGGUGCAUUUAGUACUCACUUC ..((.(((((......((((((((((....(((.((((((((.((.........)).)))))))).))).(((..((((..-..))))..))).))))))).)))....))))))).... ( -35.30) >DroSim_CAF1 174718 119 - 1 AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCAUUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUACA-AAUAUGAGCGCCAGCUGGGUGCAUUUAGUACUCACUUC ((((((((.((.((...(((.(((...)))))).))))))))))))...(((((...((((((.......(((..((((..-..))))..))))))))))))))................ ( -35.31) >DroEre_CAF1 170215 120 - 1 AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCCGCAGCUGCGUCUAAGCGCCAUUCAGCUGCAAAUUAGCGAACACACGGAGUAUGAACGCCAGCUGGGCGCAUUUAGUACUCACUUC ..((.(((((......((((((((((.........((((((((((....)))).....))))))................((.((....)).))))))))).)))....))))))).... ( -33.80) >DroYak_CAF1 171042 120 - 1 AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGCUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUACAAAAUAUGAGCGCCAGCUGGGCGCAUUUAGUACUCACUUC ..((.(((((......((((((((((....(((.(((((((((((....)))).....))))))).))).(((..((((.....))))..))).))))))).)))....))))))).... ( -40.30) >consensus AAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGUUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUACA_AAUAUGAGCGCCAGCUGGGUGCAUUUAGUACUCACUUC ..((.(((((......((((((((((....(((.((((((((.((.........)).)))))))).))).(((..((((.....))))..))).))))))).)))....))))))).... (-33.92 = -34.28 + 0.36)

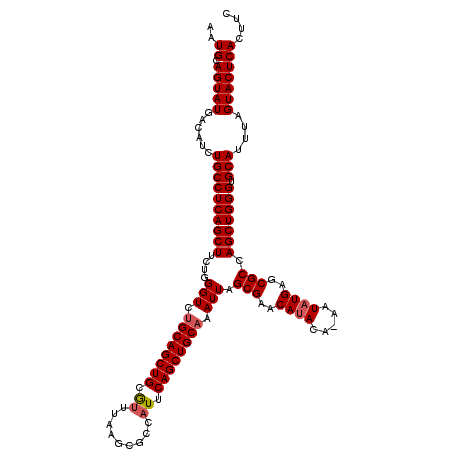
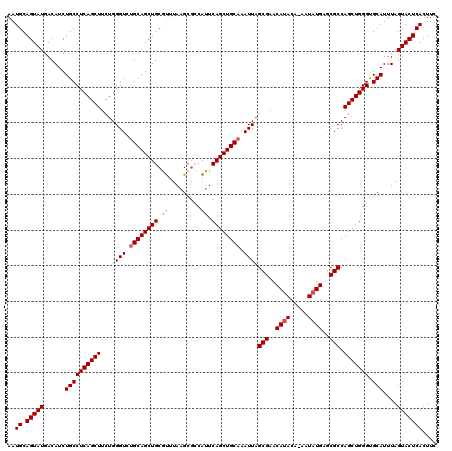
| Location | 19,989,402 – 19,989,520 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19989402 118 + 22407834 GUAUGUUCGCUAAUUUGCAGCUGAAUGGAGCUUAAACGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUGUGGUGG- .......(((((.((((((((((..(.........)..))))))))))(((((.((((....(((((((......)))))))(((.....))).)))).-.))))).......))))).- ( -40.00) >DroSec_CAF1 172187 119 + 1 GUAUGUUCGCUAAUUUGCAGCUGAAUGUCGCUUAAAUGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUCGGGUGGG ......(((((..((((((((((.((.........)).))))))))))(((((.((((....(((((((......)))))))(((.....))).)))).-.)))))........))))). ( -40.20) >DroSim_CAF1 174757 119 + 1 GUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAAUGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUCGGGUGGG ......(((((..((((((((((.((.........)).))))))))))(((((.((((....(((((((......)))))))(((.....))).)))).-.)))))........))))). ( -40.20) >DroEre_CAF1 170255 110 + 1 GUGUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAGACGCAGCUGCGGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAA-AGCAGGCUAGACUGGGUGGAA--------- (((....)))....(((((((((.....(.....)...)))))))))((((((.((((....(((((((......)))))))(((....-))).))))...))))))....--------- ( -38.10) >DroYak_CAF1 171082 111 + 1 GUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAGCGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGAAACACCAGGCUAGACUGGGAAAAA--------- .............(((((((((.....((((....)))))))))))))(((((.((((.((.(((((((......))))))).))(......).))))...))))).....--------- ( -37.60) >consensus GUAUGUUCGCUAAUUUGCAGCUGAAUGGCGCUUAAACGCAGCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA_ACUGGGGAAAAU__GGUGG_ .............((((((((((...............))))))))))(((((.((((....(((((((......)))))))(((.....))).))))...))))).............. (-33.56 = -33.60 + 0.04)

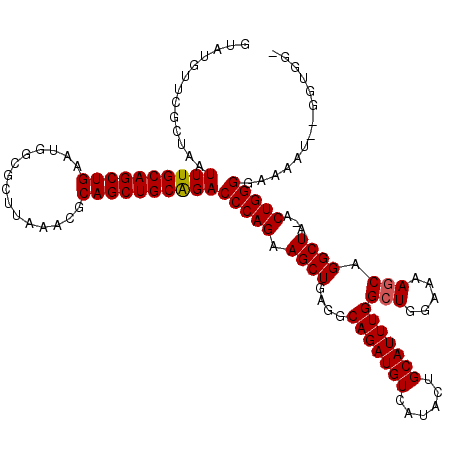
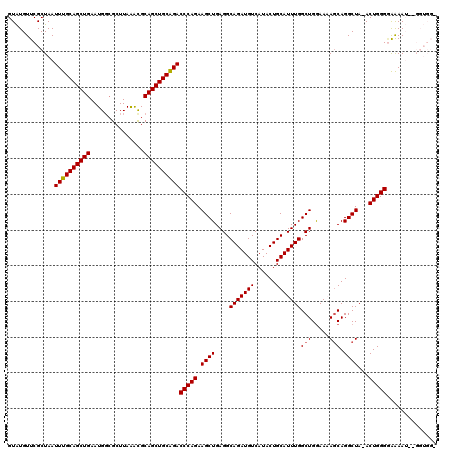
| Location | 19,989,402 – 19,989,520 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.54 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.74 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19989402 118 - 22407834 -CCACCACAUUUUCCCCAGU-UAGCCUGCUUUUCCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGUUUAAGCUCCAUUCAGCUGCAAAUUAGCGAACAUAC -.............(((((.-.(((..(((.....))).....((((........))))....))).)))))..((((((((.((.........)).))))))))............... ( -28.40) >DroSec_CAF1 172187 119 - 1 CCCACCCGAUUUUCCCCAGU-UAGCCUGCUUUUCCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCAUUUAAGCGACAUUCAGCUGCAAAUUAGCGAACAUAC ......((......(((((.-.(((..(((.....))).....((((........))))....))).)))))..((((((((.((.........)).))))))))......))....... ( -28.60) >DroSim_CAF1 174757 119 - 1 CCCACCCGAUUUUCCCCAGU-UAGCCUGCUUUUCCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCAUUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUAC ......((......(((((.-.(((..(((.....))).....((((........))))....))).)))))..((((((((.((.........)).))))))))......))....... ( -28.60) >DroEre_CAF1 170255 110 - 1 ---------UUCCACCCAGUCUAGCCUGCU-UUCCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCCGCAGCUGCGUCUAAGCGCCAUUCAGCUGCAAAUUAGCGAACACAC ---------((((((((((...(((..(((-....))).....((((........))))....))).))))))..((((((((((....)))).....))))))......).)))..... ( -31.50) >DroYak_CAF1 171082 111 - 1 ---------UUUUUCCCAGUCUAGCCUGGUGUUUCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGCUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUAC ---------.....(((((...(((.((((......))))...((((........))))....))).)))))..(((((((((((....)))).....)))))))............... ( -35.80) >consensus _CCACC__AUUUUCCCCAGU_UAGCCUGCUUUUCCAGCCAAAUGCAGUAUGACAUCUGCCUCAGCUUCUGGGUCUGCAGCUGCGUUUAAGCGCCAUUCAGCUGCAAAUUAGCGAACAUAC ..............(((((...((((((......)))......((((........))))....))).)))))...(((((((.((.........)).)))))))................ (-26.78 = -26.74 + -0.04)

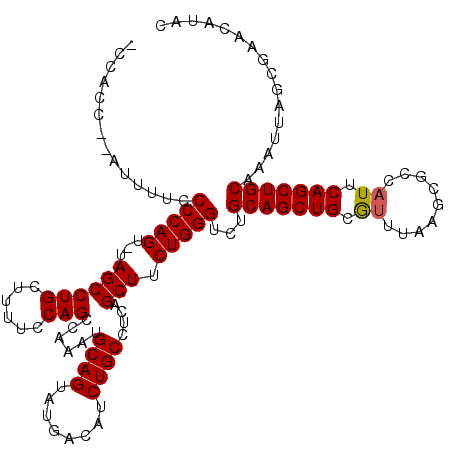
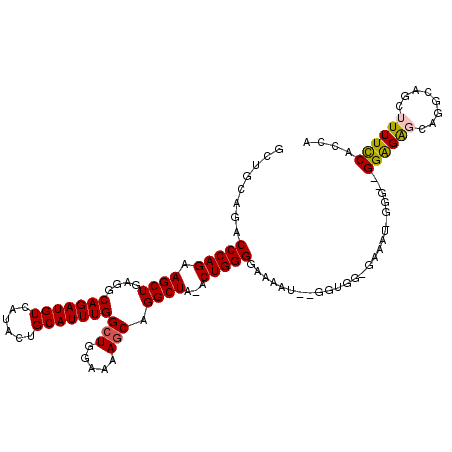
| Location | 19,989,442 – 19,989,555 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.36 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19989442 113 + 22407834 GCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUGUGGUGG-GAAAUGGGGGAGGGGAGGAGGUAGCUUUUCCACCA ((..((..(((((.((((....(((((((......)))))))(((.....))).)))).-.))))).....))..))..-...........(((((((((....))))))).)). ( -37.60) >DroSec_CAF1 172227 112 + 1 GCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUCGGGUGGGGAAAUACGG--GGAGAGGAGGCAGCUUUUCCACCA .(..(.(((((((.((((....(((((((......)))))))(((.....))).)))).-.))))).....))..)..)........((--(((((((......))))))).)). ( -34.30) >DroSim_CAF1 174797 112 + 1 GCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA-ACUGGGGAAAAUCGGGUGGGGAAAUGGGG--GGAGAGCAGGCAGCUUUUCCACCA .(..(.(((((((.((((....(((((((......)))))))(((.....))).)))).-.))))).....))..)..).....(((.(--(((((((.....)))))))).))) ( -37.50) >DroEre_CAF1 170295 95 + 1 GCUGCGGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAA-AGCAGGCUAGACUGGGUGGAA---------------AGG--GGAGG--AGGCUGCUUUUCCCCCA .......((((((.((((....(((((((......)))))))(((....-))).))))...))))))....---------------.((--(((((--((....))))))))).. ( -37.60) >DroYak_CAF1 171122 98 + 1 GCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGAAACACCAGGCUAGACUGGGAAAAA---------------GGG--GGAGACCAGGCAAUCUUUUCACCA (((......((...(((..((((((........)))).))..)))......((((......))))......---------------))(--(....)).)))............. ( -27.90) >consensus GCUGCAGACCCAGAAGCUGAGGCAGAUGUCAUACUGCAUUUGGCUGGAAAAGCAGGCUA_ACUGGGGAAAAU__GGUGG_GAAAU_GGG__GGAGAGCAGGCAGCUUUUCCACCA ........(((((.((((....(((((((......)))))))(((.....))).))))...))))).........................((((((........)))))).... (-23.24 = -23.36 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:16 2006