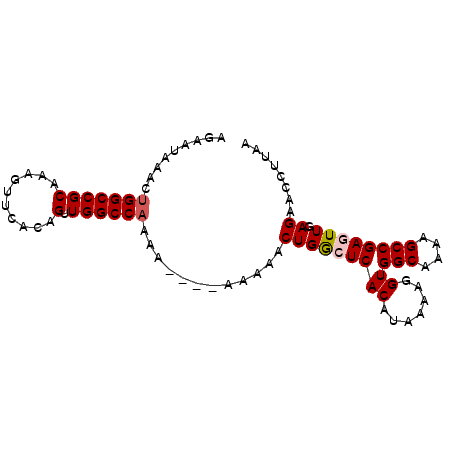
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,983,018 – 19,983,108 |
| Length | 90 |
| Max. P | 0.822017 |

| Location | 19,983,018 – 19,983,108 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -13.74 |
| Energy contribution | -14.34 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19983018 90 + 22407834 UUAAGGUUCUCAACUCGGCUUUUGCCACCUUUUAUGUGAGCCAGUUUUUUUCUUUUUGGCCAACUGUGAACUUUGCGGCCAGUUCAUUCU .((((((((.((....(((....))).........((..(((((...........)))))..)))).))))))))............... ( -20.00) >DroSec_CAF1 165899 86 + 1 UUAAGGUUCUCAACUCGGCUUUUGCCACCUUUUAUGUGAGCCAGUUUUU----UUUUGGCCAACUGUGAACUUUGCGGCCAGUUUAUUCU .((((((((.((....(((....))).........((..(((((.....----..)))))..)))).))))))))............... ( -20.70) >DroSim_CAF1 168435 86 + 1 UUAAGGUUCUCAACUCGGCUUUUGCCACCUUUUAUGUGAGCCAGUUUUU----UUUUGGCCAACUGUGAACUUUGCGGCCAGUUUAUUCU .((((((((.((....(((....))).........((..(((((.....----..)))))..)))).))))))))............... ( -20.70) >DroEre_CAF1 163941 77 + 1 -----AUUCUCAACUCGGCUUUUGCCACCUUUU-GGUGACUCAGUUU-------UUGGGCCAACUGUGAACUUUGCGGCCAGUCUGUUCU -----...........(((....)))(((....-)))((..(((...-------((((.((((..(....).))).).)))).))).)). ( -17.50) >DroYak_CAF1 164842 78 + 1 -----GUUCUUAACUCGGCUUUUGCCACCUUUUCGGUGACUCAGUUU-------UUUGGCCAACUGUGAACUUUGCGGCCAGUUUAUUCU -----......((((.((((...((((((.....)))).(.(((((.-------.......))))).)......))))))))))...... ( -16.50) >consensus UUAAGGUUCUCAACUCGGCUUUUGCCACCUUUUAUGUGAGCCAGUUUUU____UUUUGGCCAACUGUGAACUUUGCGGCCAGUUUAUUCU ....(((((.......(((....)))((.......))))))).............((((((...............))))))........ (-13.74 = -14.34 + 0.60)

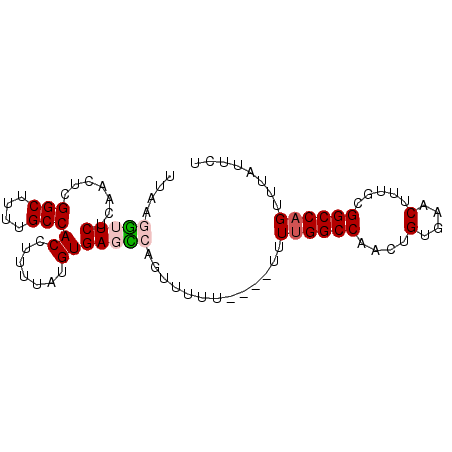
| Location | 19,983,018 – 19,983,108 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -18.92 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.38 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19983018 90 - 22407834 AGAAUGAACUGGCCGCAAAGUUCACAGUUGGCCAAAAAGAAAAAAACUGGCUCACAUAAAAGGUGGCAAAAGCCGAGUUGAGAACCUUAA ....((((((........))))))..((.(((((.............))))).))....((((((((....))).........))))).. ( -19.12) >DroSec_CAF1 165899 86 - 1 AGAAUAAACUGGCCGCAAAGUUCACAGUUGGCCAAAA----AAAAACUGGCUCACAUAAAAGGUGGCAAAAGCCGAGUUGAGAACCUUAA ...................((((.((((.(((((...----......))))).))........((((....))))...)).))))..... ( -16.00) >DroSim_CAF1 168435 86 - 1 AGAAUAAACUGGCCGCAAAGUUCACAGUUGGCCAAAA----AAAAACUGGCUCACAUAAAAGGUGGCAAAAGCCGAGUUGAGAACCUUAA ...................((((.((((.(((((...----......))))).))........((((....))))...)).))))..... ( -16.00) >DroEre_CAF1 163941 77 - 1 AGAACAGACUGGCCGCAAAGUUCACAGUUGGCCCAA-------AAACUGAGUCACC-AAAAGGUGGCAAAAGCCGAGUUGAGAAU----- ......(((((((...........(((((.......-------.))))).((((((-....))))))....))).))))......----- ( -21.60) >DroYak_CAF1 164842 78 - 1 AGAAUAAACUGGCCGCAAAGUUCACAGUUGGCCAAA-------AAACUGAGUCACCGAAAAGGUGGCAAAAGCCGAGUUAAGAAC----- ......(((((((...........(((((.......-------.))))).((((((.....))))))....))).))))......----- ( -21.90) >consensus AGAAUAAACUGGCCGCAAAGUUCACAGUUGGCCAAAA____AAAAACUGGCUCACAUAAAAGGUGGCAAAAGCCGAGUUGAGAACCUUAA .........(((((((..........).))))))............(((((((((.......))(((....)))))))).))........ (-14.02 = -14.38 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:09 2006