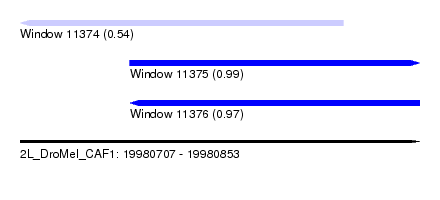
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,980,707 – 19,980,853 |
| Length | 146 |
| Max. P | 0.991362 |

| Location | 19,980,707 – 19,980,825 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.92 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19980707 118 - 22407834 AUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUCAGAAUUGUGUGAUGCCGGGCAGUGAUGUGGGGCAGAAACUUAAAAA ....((((((((..(((((((....)))))))((((((.....((((((...........)))))))))))).))).)))))..((((..(((....)))..))))............ ( -29.30) >DroSec_CAF1 163611 118 - 1 AUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUUGUGUGAUGCCGGGCAGUGAUGUGGGGCAGAAACUUAAAAA .(((((((((.........((((((((((((.(.(((((((........))))))).).)))).....))))))))..))))).((((..(((....)))..))))....)))).... ( -28.00) >DroSim_CAF1 166137 117 - 1 AUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAAGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUUGUGUGAUGCCGGGCAGUGAUGUGGGGCAGAAACUUAAAA- .(((((((((.........((((((((.((((...(((((((.......)))))))......))))..))))))))..))))).((((..(((....)))..))))....))))...- ( -27.00) >DroEre_CAF1 161750 106 - 1 AUGGGCUCAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUUGUGUGAUGCCG------------UGGCAGAAACUUAAAAA ...(((((((.((......((((((((((((.(.(((((((........))))))).).)))).....)))))))).)).)))).))).------------................. ( -23.00) >DroYak_CAF1 162622 106 - 1 AUGGGCUCAUUCGUUUUAUUUCUCAGAAUAAGAGGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUUGUGUGAUGCCG------------GGGCAGAAACUUAAAAA ....((((...((((.((.((((((((((((.(.(((((((........))))))).).)))).....)))))))).))...))))...------------))))............. ( -23.10) >consensus AUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUUGUGUGAUGCCGGGCAGUGAUGUGGGGCAGAAACUUAAAAA .((((.....((((..((.(((((((((((((((((.............)))))))))((.....)).)))))))).)).))))((((..(((....)))..))))....)))).... (-21.78 = -21.80 + 0.02)

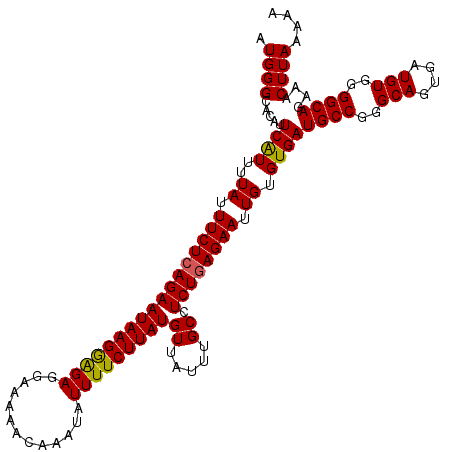
| Location | 19,980,747 – 19,980,853 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19980747 106 + 22407834 AAUUCUGAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCCUCUCCUUAUUCUGAGAAAUAAAAUGAAUGUGCCCAUUAGCUUGUGCAAACAGACAGGUGGGGAA .((((.((((((((((((...........)))))))))))).((((........))))........))))...(((((..(..(((....)))..)..)))))... ( -23.80) >DroSec_CAF1 163651 106 + 1 AAUUCUCAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCCUCUCCUUAUUCUGAGAAAUAAAAUGAAUGUGCCCAUUAGCUUGUGCAAACAGACAGGUGGGGCA ..((((((((((((((((...........)))))))..............)))))))))............(((((....((((((........)))))).))))) ( -26.43) >DroSim_CAF1 166176 106 + 1 AAUUCUCAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCUUCUCCUUAUUCUGAGAAAUAAAAUGAAUGUGCCCAUUAGCUUGUGUAAACAGACAGGUGGGGCA ..(((((((((...........(((((((.......))))))).......)))))))))............(((((....((((((........)))))).))))) ( -28.07) >DroEre_CAF1 161778 106 + 1 AAUUCUCAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCCUCUCCUUAUUCUGAGAAAUAAAAUGAAUGAGCCCAUUAGCUUGUGCAAACAGACAGGUGGGGAA ........((((((((((...........))))))))))(((((.(((...(((((....)....((.(..(((......)))..).))..)))).))).))))). ( -23.10) >DroYak_CAF1 162650 106 + 1 AAUUCUCAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCCUCCUCUUAUUCUGAGAAAUAAAACGAAUGAGCCCAUUAGCUUGUGCAAACAGACAGGUGGGGAA ..((((((((((((((((...........)))))))..............)))))))))..............(((((..(..(((....)))..)..)))))... ( -22.93) >consensus AAUUCUCAGAGGCAAAUAACAUAAGAAAAUAUUUGUUUUUCCUCUCCUUAUUCUGAGAAAUAAAAUGAAUGUGCCCAUUAGCUUGUGCAAACAGACAGGUGGGGAA ..((((((((((((((((...........)))))))..............)))))))))..............(((((..(..(((....)))..)..)))))... (-21.83 = -22.03 + 0.20)

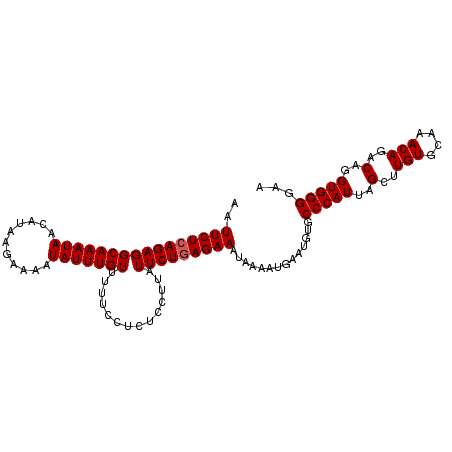
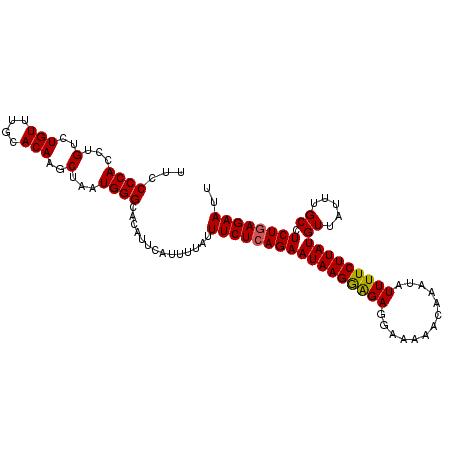
| Location | 19,980,747 – 19,980,853 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19980747 106 - 22407834 UUCCCCACCUGUCUGUUUGCACAAGCUAAUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUCAGAAUU .........((((((((.((....)).)))))))).((((..(((((((....)))))))((((((.....((((((...........)))))))))))).)))). ( -22.10) >DroSec_CAF1 163651 106 - 1 UGCCCCACCUGUCUGUUUGCACAAGCUAAUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUU (((((.....(..(((....)))..)....)))))............((((((((((((.(.(((((((........))))))).).)))).....)))))))).. ( -24.30) >DroSim_CAF1 166176 106 - 1 UGCCCCACCUGUCUGUUUACACAAGCUAAUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAAGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUU (((((.....(..(((....)))..)....)))))............((((((((.((((...(((((((.......)))))))......))))..)))))))).. ( -23.30) >DroEre_CAF1 161778 106 - 1 UUCCCCACCUGUCUGUUUGCACAAGCUAAUGGGCUCAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUU ..........(((((((.((....)).))))))).............((((((((((((.(.(((((((........))))))).).)))).....)))))))).. ( -21.90) >DroYak_CAF1 162650 106 - 1 UUCCCCACCUGUCUGUUUGCACAAGCUAAUGGGCUCAUUCGUUUUAUUUCUCAGAAUAAGAGGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUU ..........(((((((.((....)).))))))).............((((((((((((.(.(((((((........))))))).).)))).....)))))))).. ( -21.90) >consensus UUCCCCACCUGUCUGUUUGCACAAGCUAAUGGGCACAUUCAUUUUAUUUCUCAGAAUAAGGAGAGGAAAAACAAAUAUUUUCUUAUGUUAUUUGCCUCUGAGAAUU ...((((...(..(((....)))..)...))))..............(((((((((((((((((.............)))))))))((.....)).)))))))).. (-20.40 = -20.28 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:06 2006