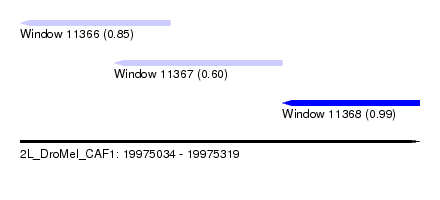
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,975,034 – 19,975,319 |
| Length | 285 |
| Max. P | 0.994658 |

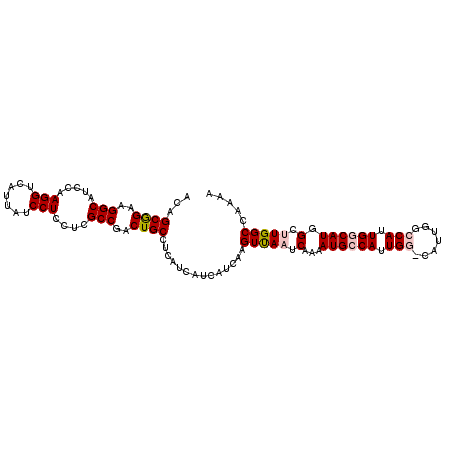
| Location | 19,975,034 – 19,975,141 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19975034 107 - 22407834 ACAGCGGAAGGCAUCCAAGGUCAUUAUCCUCCUCGCCGACUGCCUCAUCAUCAUCAAGUUAAUCAAAAUGCCAUUGGACAUUGGCCAUUGGCAUGGCUUGGCCAAAA ...((((..(((.....(((.......)))....)))..))))..........(((((((.......((((((.(((.......))).)))))))))))))...... ( -28.11) >DroSec_CAF1 157914 107 - 1 ACAGCGGAAGGCAUCCCAGGUCAUUAUCCUCCUCGCCGACUGCCUCAUCAUCAUCAAGUUAAUCAAAAUGCCAUUGGCCAUUGGCCAUUGGCAUGGCUUGGCCAAAA ...((((..(((.....(((.......)))....)))..))))..........(((((((.......((((((.(((((...))))).)))))))))))))...... ( -33.71) >DroSim_CAF1 160433 107 - 1 ACAGCGGAAGGCAUCCCAGGUCAUUAUCCUCCUCGCCGACUGCCUCAUCAUCAUCAAGUUAAUCAAAAUGCCAUUGGCCAUUGGCCAUUGGCAUGGCUUGGCCAAAA ...((((..(((.....(((.......)))....)))..))))..........(((((((.......((((((.(((((...))))).)))))))))))))...... ( -33.71) >DroEre_CAF1 155980 100 - 1 ACAGCGGAAGGCACCCGAGGUCAUUAUCCUCCUCGCCGACUGCCUCAUCAUCAUCAAGUUAAUCAAAAUGCCAUUG-------GCCAUUGCCAUGGCAUGGCCAAAA ...((((..(((....((((.......))))...)))..))))..............................(((-------(((((.((....)))))))))).. ( -31.30) >DroYak_CAF1 156927 100 - 1 ACAGCAGAAGGCACCCAAGGUCAUUAUCCUCCUCGCCAACUGCCUCAUCAUCAUCGAGUCAAUCAAAAUGCCAUUG-------GCCAUUGCCAUGGCAUGGCCGAAA ...((((..(((.....(((.......)))....)))..))))..........(((.((((.......((((((.(-------((....)))))))))))))))).. ( -29.81) >consensus ACAGCGGAAGGCAUCCAAGGUCAUUAUCCUCCUCGCCGACUGCCUCAUCAUCAUCAAGUUAAUCAAAAUGCCAUUGG_CAUUGGCCAUUGGCAUGGCUUGGCCAAAA ...((((..(((.....(((.......)))....)))..))))..............(((((.(...((((((.(((.......))).)))))).).)))))..... (-22.50 = -23.38 + 0.88)

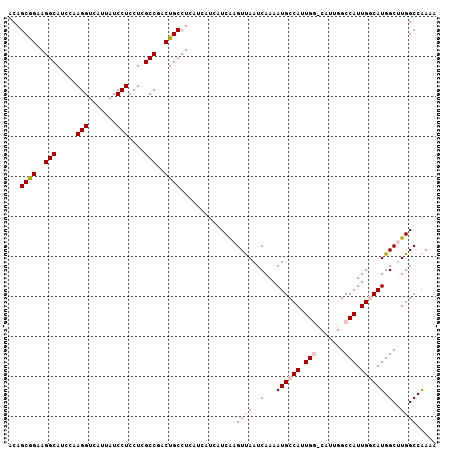
| Location | 19,975,101 – 19,975,221 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -35.02 |
| Energy contribution | -34.14 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19975101 120 - 22407834 GUUGGCCAUCUAACCGCAAUCAGGAGAUCGAAGGAAAUGCCACUUCGGAGGAAACGAGGAUGAGCCAACUGCCCACGUUGACAGCGGAAGGCAUCCAAGGUCAUUAUCCUCCUCGCCGAC ((((((.((((..((.......))))))(((((.........)))))(((((.....(((((((((...((((..(((.....)))...)))).....)))..))))))))))))))))) ( -36.70) >DroSec_CAF1 157981 120 - 1 GUUGGCCAUUUAACCGCAAUCAGGAGAUCGAAGGAAAUGCCACUUCGGAGGAAACGAGGAUGAGCCAACUGCCCACGUUGACAGCGGAAGGCAUCCCAGGUCAUUAUCCUCCUCGCCGAC ((((((.......((.......)).((((((((.........)))))).(....)(((((((((((...((((..(((.....)))...)))).....)))..)))))))).)))))))) ( -36.60) >DroSim_CAF1 160500 120 - 1 GUUGGCCAUCUAACCGCAAUCGGGAGAUCGAAGGAAAUGCCACUUCGGAGGAAACGAGGAUGAGCCAACUGCCCACGUUGACAGCGGAAGGCAUCCCAGGUCAUUAUCCUCCUCGCCGAC ((((((.......(((....)))(((.((((((.........)))))).(....)(((((((((((...((((..(((.....)))...)))).....)))..))))))))))))))))) ( -40.90) >DroEre_CAF1 156040 119 - 1 GUUGGCCAUCUAACCGCAAUCAGGAGAUCGAAGGAAAUGCCACUUCGGAGGAAGCGAGGAUGAGCCAACUGC-CACAUUGACAGCGGAAGGCACCCGAGGUCAUUAUCCUCCUCGCCGAC ((((((..(((..((.......))...((((((.........)))))).)))((.(((((((((((..((((-..........))))..)))(((...)))..)))))))))).)))))) ( -36.50) >DroYak_CAF1 156987 120 - 1 GUUGGCCAUCCAACCACAAUCAGGAGAUCGAAGGAAAUGCCACUUCGGUGGAAACGAGGAUGAGCCAACUGCCCACAUGGACAGCAGAAGGCACCCAAGGUCAUUAUCCUCCUCGCCAAC ((((((..(((..((.......))..(((((((.........))))))))))...(((((((((((..((((((....))...))))..)))(((...)))..))))))))...)))))) ( -42.00) >consensus GUUGGCCAUCUAACCGCAAUCAGGAGAUCGAAGGAAAUGCCACUUCGGAGGAAACGAGGAUGAGCCAACUGCCCACGUUGACAGCGGAAGGCAUCCAAGGUCAUUAUCCUCCUCGCCGAC ((((((..(((..((.......))...((((((.........)))))).)))...(((((((((((..((((...........))))..)))((.....))..))))))))...)))))) (-35.02 = -34.14 + -0.88)

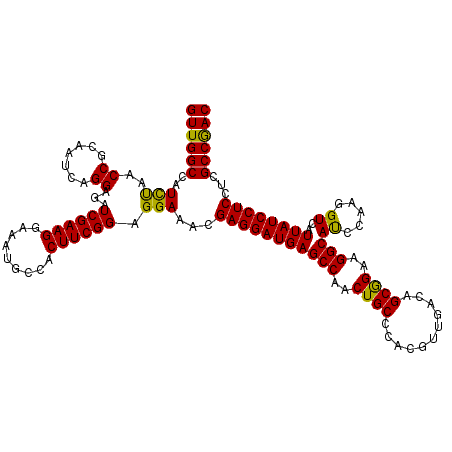
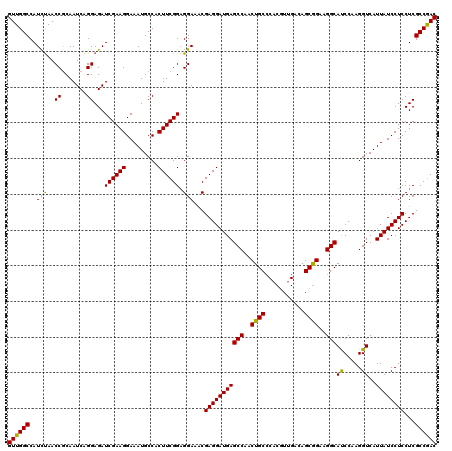
| Location | 19,975,221 – 19,975,319 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -32.36 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19975221 98 - 22407834 GUUGGCUUGGCUGAGAGGCUCAUGGCCUCGCCAAAAGCGCAGUGAGCCGGAAUUUGCUUUCUCGGCCACGUUUAGUUCGAGGGGAUCUGGCUUAUUAA ....(((((((...(((((.....)))))))))..)))..(((((((((((......(..((((..(.......)..))))..).))))))))))).. ( -37.30) >DroSec_CAF1 158101 98 - 1 GUUGGCUUGGCUGAGAAGCUCUUGGCUUCGCCAAAAGCCCAGUGAGCCGGAAUCCGCUUUCUCGGCCACGUUUAGUUCGAGGGGAUCUGGCUUAUUAA .(((((..(((..((.....))..)))..)))))......(((((((((((.(((....(((((..(.......)..))))))))))))))))))).. ( -40.50) >DroSim_CAF1 160620 98 - 1 GUUGGCUUGGCUGAGAAGCUCUUGGCUUCGCCAAAAGCGCAGUGAGCCGGAAUCCGCUUUCUCGGCCACGUUUAGUUCGAGGGGAUCUGGCUUAUUAA .(((((..(((..((.....))..)))..)))))......(((((((((((.(((....(((((..(.......)..))))))))))))))))))).. ( -40.50) >DroEre_CAF1 156159 97 - 1 GUUGGCUUGACGCU-CAGCUCAUGGCUUCGCCAAAAGCGCAGUGAGCCGGAAUCCGCUUUCUCGGCCACGUUUAGUUUGGGGGGAUCUGGCUUAUUAA ..((((.....)).-))(((..((((...))))..)))..(((((((((((.(((.(((.((..((...))..))...))).)))))))))))))).. ( -32.50) >DroYak_CAF1 157107 97 - 1 GUUGGCUUGACUCU-CAGCUCAUGGCUUCGCCAAAAGCGCAGUGAGCCGGAAUUCGCUUUCUCGGCCACGUUUAGUUUGAGGGGAUCUGGCUUAUUAA ...((((.((((((-(((((..((((...))))..)))...(((.(((((((......))).)))))))........))))))..)).))))...... ( -33.00) >consensus GUUGGCUUGGCUGAGAAGCUCAUGGCUUCGCCAAAAGCGCAGUGAGCCGGAAUCCGCUUUCUCGGCCACGUUUAGUUCGAGGGGAUCUGGCUUAUUAA .(((((..(((((((.....)))))))..)))))......(((((((((((.(((.(((.((..((...))..))...))).)))))))))))))).. (-32.36 = -32.92 + 0.56)

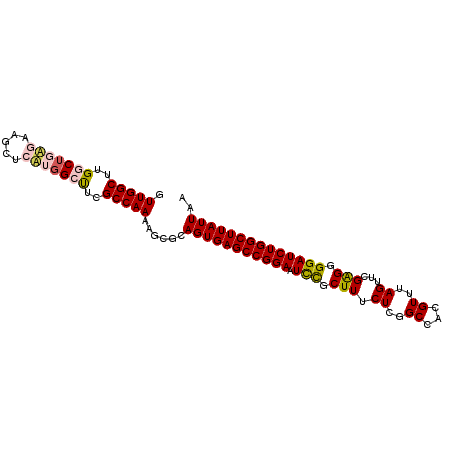
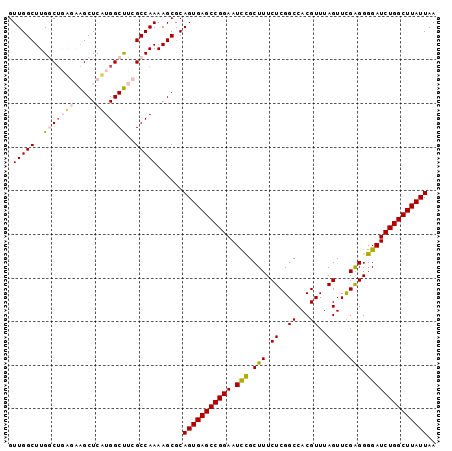
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:59 2006