
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,072,843 – 2,072,934 |
| Length | 91 |
| Max. P | 0.964390 |

| Location | 2,072,843 – 2,072,934 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -42.08 |
| Consensus MFE | -31.41 |
| Energy contribution | -32.50 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2072843 91 + 22407834 AAGGCAGGACUAAAACCAGGACCUGCAACCUGGCAGCUCCUGGCU-------CCUGCCAGCGCUGCAGGCCGGGAUUAGGUUCAGGGCGGGUCCUUGC-- ...(((((.......))(((((((((..(((((((((..(((((.-------...))))).))))).((((.......))))))))))))))))))))-- ( -44.00) >DroSec_CAF1 89857 91 + 1 AAGGCAGGACUAAAACCAGGACCUGCAACCUGGCAGCUCCUGGCU-------CCUGCCAGCGCUGCAGGCCGGGAUUAGGUUCAGGGCGGGUCCUUGC-- ...(((((.......))(((((((((..(((((((((..(((((.-------...))))).))))).((((.......))))))))))))))))))))-- ( -44.00) >DroSim_CAF1 89178 91 + 1 AAGGCAGGACUAAAACCAGGACCUGCAACCUGGCAGCUCCUGGCU-------CCUGCCAGCGCUGCAGGCCGGGAUUAGGUUCAGGGCGGGUCCUUGC-- ...(((((.......))(((((((((..(((((((((..(((((.-------...))))).))))).((((.......))))))))))))))))))))-- ( -44.00) >DroEre_CAF1 92255 98 + 1 AAGCCAGUGCUAAGAGCAGGACCUGCAACCUAUCAGCUCCUGGCUCCUUGCUCCUGCCAGAGCUGCAGGCCAGGAUUAGGUUCAGGGCGGGUCCUUGC-- .(((....)))....(((((((((((((((((.....((((((((....((((......))))....)))))))).))))))....)))))))).)))-- ( -44.20) >DroYak_CAF1 93614 91 + 1 AAGGCAGGACUAAAACCAGGACCUGCAACCUGGCAGCUCCUGGCU-------GCUGCCAGAGCUGCAGGCCAGGAUUAGGUUCAGGGCAGGUCCUUGC-- ...(((((.......))(((((((((..(((((((((((.((((.-------...))))))))))).((((.......))))))))))))))))))))-- ( -46.60) >DroAna_CAF1 89066 80 + 1 ---CCAGGAGCAUCGCCAGGACAUGCAACCUGGCAGCUCC--------------AACGAUUGCUGCAAGCUAAGAUUAGCUGCAU---AUGUCCUUCUCC ---...(((((...((((((........)))))).)))))--------------...(((...((((.((((....)))))))).---..)))....... ( -29.70) >consensus AAGGCAGGACUAAAACCAGGACCUGCAACCUGGCAGCUCCUGGCU_______CCUGCCAGCGCUGCAGGCCAGGAUUAGGUUCAGGGCGGGUCCUUGC__ ......((.......))(((((((((..(((((((((..(((((...........))))).)))))..(((.......))).)))))))))))))..... (-31.41 = -32.50 + 1.09)

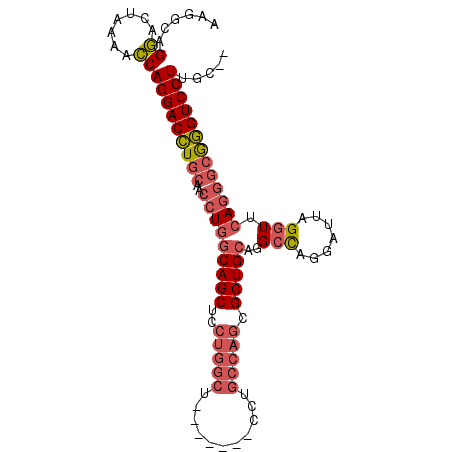
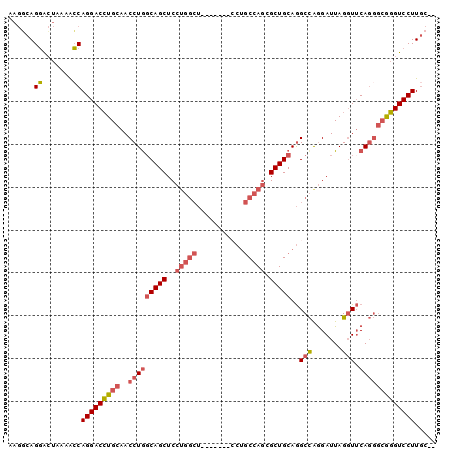
| Location | 2,072,843 – 2,072,934 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2072843 91 - 22407834 --GCAAGGACCCGCCCUGAACCUAAUCCCGGCCUGCAGCGCUGGCAGG-------AGCCAGGAGCUGCCAGGUUGCAGGUCCUGGUUUUAGUCCUGCCUU --(((.((((.......(((((.......((((((((((.(((((((.-------.........)))))))))))))))))..)))))..)))))))... ( -40.70) >DroSec_CAF1 89857 91 - 1 --GCAAGGACCCGCCCUGAACCUAAUCCCGGCCUGCAGCGCUGGCAGG-------AGCCAGGAGCUGCCAGGUUGCAGGUCCUGGUUUUAGUCCUGCCUU --(((.((((.......(((((.......((((((((((.(((((((.-------.........)))))))))))))))))..)))))..)))))))... ( -40.70) >DroSim_CAF1 89178 91 - 1 --GCAAGGACCCGCCCUGAACCUAAUCCCGGCCUGCAGCGCUGGCAGG-------AGCCAGGAGCUGCCAGGUUGCAGGUCCUGGUUUUAGUCCUGCCUU --(((.((((.......(((((.......((((((((((.(((((((.-------.........)))))))))))))))))..)))))..)))))))... ( -40.70) >DroEre_CAF1 92255 98 - 1 --GCAAGGACCCGCCCUGAACCUAAUCCUGGCCUGCAGCUCUGGCAGGAGCAAGGAGCCAGGAGCUGAUAGGUUGCAGGUCCUGCUCUUAGCACUGGCUU --(((.(((((.((....((((((.(((((((((...(((((....)))))..)).))))))).....)))))))).))))))))....(((....))). ( -43.20) >DroYak_CAF1 93614 91 - 1 --GCAAGGACCUGCCCUGAACCUAAUCCUGGCCUGCAGCUCUGGCAGC-------AGCCAGGAGCUGCCAGGUUGCAGGUCCUGGUUUUAGUCCUGCCUU --(((.((((.......(((((.......((((((((((.((((((((-------........))))))))))))))))))..)))))..)))))))... ( -45.10) >DroAna_CAF1 89066 80 - 1 GGAGAAGGACAU---AUGCAGCUAAUCUUAGCUUGCAGCAAUCGUU--------------GGAGCUGCCAGGUUGCAUGUCCUGGCGAUGCUCCUGG--- ((((.(((((((---.((((((((....)))).))))((((((..(--------------((.....)))))))))))))))).......))))...--- ( -31.30) >consensus __GCAAGGACCCGCCCUGAACCUAAUCCCGGCCUGCAGCGCUGGCAGG_______AGCCAGGAGCUGCCAGGUUGCAGGUCCUGGUUUUAGUCCUGCCUU .....((((((.((...............((((((((((.(((((...........)))))..)))).)))))))).))))))(((.........))).. (-27.48 = -27.93 + 0.45)

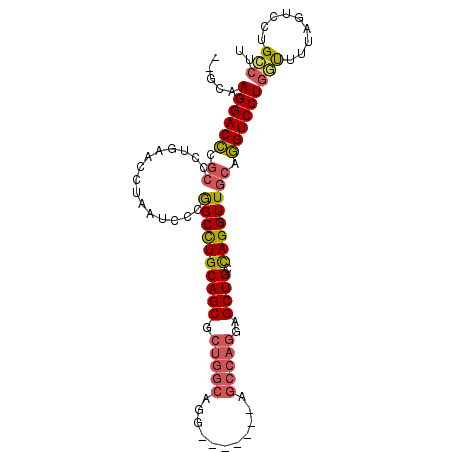
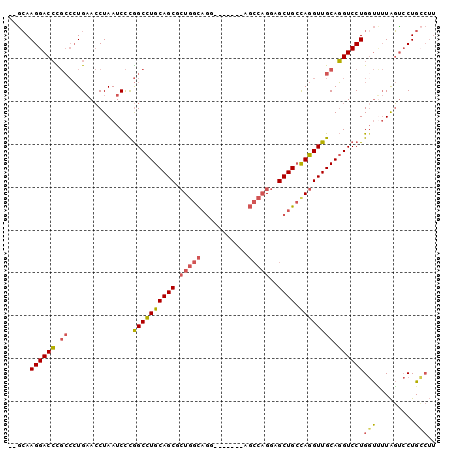
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:36 2006