
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
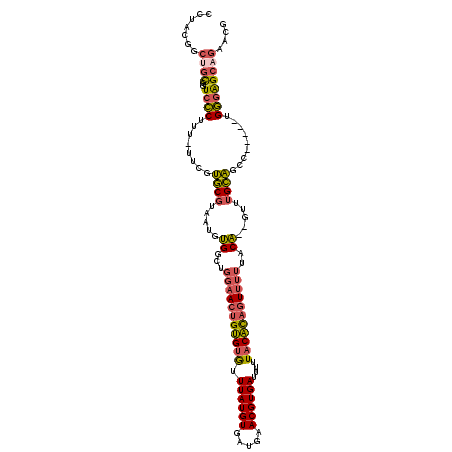
| Location | 19,970,127 – 19,970,236 |
| Length | 109 |
| Max. P | 0.670455 |

| Location | 19,970,127 – 19,970,236 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.56 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
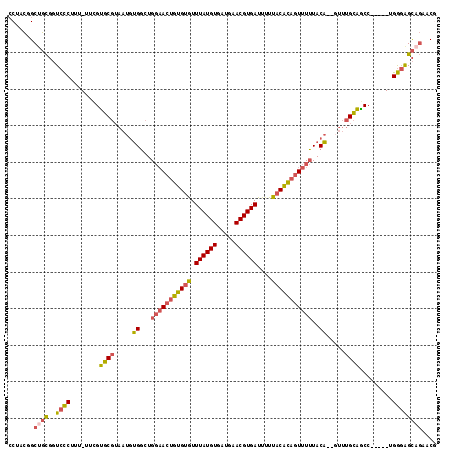
>2L_DroMel_CAF1 19970127 109 + 22407834 CCUACGGCUGCGGUCCCUUU-UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGAACGUGAUUUUUACACAGUUUUUACA--GUUUGCAGCC-----UGGGAGCAGAACG ((((.((((((((.......-..(((.....)))..((((((((((((((.((((((.....))))))....))))))))))...))--))))))))))-----))))......... ( -35.90) >DroSec_CAF1 153046 109 + 1 CCUACAGCUGCGGUCCCUUU-UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGAACGUGAUUUUUACACAGUUUUUACA--GUUUGCAGCC-----UGGAAGCAGAACG .......((((..(((....-..(((.....))).(((((((((((((((.((((((.....))))))....)))))))))).....--.....)))))-----.))).)))).... ( -31.50) >DroSim_CAF1 155621 109 + 1 CCUACAGCUGCGGUCCCUUU-UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGAACGUGAUUUUUACACAGUUUUUACA--GUUUGCAGCC-----UGGGAGCAGAACG .......((((..((((...-....((((...(((((...((((((((((.((((((.....))))))....)))))))))))))))--...))))...-----.)))))))).... ( -33.30) >DroEre_CAF1 151332 109 + 1 CCUACGGCUGCGGUCCCUUU-UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGCACGUGAUUUUUACACAGUUUUUACA--GUUUGCAACC-----AGGGAGCAGAGCU .....((((..(.(((((..-....((((...(((((...((((((((((.(((((((...)))))))....)))))))))))))))--...))))...-----))))).)..)))) ( -37.00) >DroWil_CAF1 240642 111 + 1 -CCAACGCUUUUCUCUCUUUAUUUGCCC----UCAGUCUGC-AAUGGGGAUUUAUGUGUUAAACGUGAUUUUUACCUUGUUUAUACUUGCUCUGGGGCCUACCUUGAGGGUCCAACG -....((.....(((((.......((((----.(((...((-((..(((..((((((.....))))))......)))..).......))).))))))).......))))).....)) ( -26.05) >DroYak_CAF1 152023 109 + 1 CCUUCGGCGGCGGUCCCUUU-UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGAACGUGAUUUUUACACAGUUUUUACA--CUUUGCAGCC-----AGGGAGCAGAGCA ((((.(((.((((.......-.))))..((((.(((...(((((((((((.((((((.....))))))....)))))))))))..))--).)))).)))-----))))......... ( -36.70) >consensus CCUACGGCUGCGGUCCCUUU_UUCGUGCGUAAUGUGGCUGGAACUGUGUGUUUAUGUGAUGAACGUGAUUUUUACACAGUUUUUACA__GUUUGCAGCC_____UGGGAGCAGAACG .......((((..((((........((((.....((...(((((((((((.((((((.....))))))....)))))))))))..)).....)))).........)))))))).... (-26.50 = -26.56 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:52 2006