
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,952,563 – 19,952,673 |
| Length | 110 |
| Max. P | 0.614749 |

| Location | 19,952,563 – 19,952,673 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -37.61 |
| Consensus MFE | -25.43 |
| Energy contribution | -26.18 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19952563 110 - 22407834 ---UCCAUCAU--CAA-----AACAUACCCAUCGGAAAUUGUGGUGCAGCUGCAGGUUGGUGGUGUUGCAAGUGGCCUGUGACAUGGUGCCAAUGAUGCAGGGCAGCUUGUUGGCGGGCC ---..((((((--(((-----......((....))...(((..(.....)..))).)))))))))........(((((((((((.(.((((..........)))).).)))).))))))) ( -35.40) >DroVir_CAF1 204759 99 - 1 ---------------------AACUUACCCAGCGGAAGUUGUGAUGCAGCUGCAGAUUGGUUGUGUUGCAGGUGGCCUGCGACAUGGUGCCAAUGAUGCAGGGCAGCUUGGUGGCCGGCC ---------------------...(((((((((....))))...(((..(((((.((((((.((((((((((...))))))))))...))))))..))))).)))....)))))...... ( -41.80) >DroGri_CAF1 176706 115 - 1 AAGUUCAUCAAAGUUU-----CACUUACCCAUCGGAAAUUGUGAUGUAGCUGAAGAUUGGUCGUGUUGCAGGUGGCUUGAGACAUGGUGCCAAUGAUGCAGGGCAGCUUGGUGGCUGGGC .(((((((((.(((((-----(............)))))).))))).))))....(((((((((((..((((...))))..)))))..)))))).........(((((....)))))... ( -31.30) >DroWil_CAF1 214168 108 - 1 ---UUGUAGU----UG-----GACUUACCCAACGGAAAUUGUGAUGCAAUUGGAGAUUGGUUGUAUUGCAGGUGGCCUGGGACAUGAUGCCAAUGAUGCAUGGCAAUUUAUUGGGUGGCC ---.....((----((-----(......)))))......((..(((((((..(...)..)))))))..))(((.(((..(.......(((((........))))).....)..))).))) ( -36.00) >DroMoj_CAF1 290204 105 - 1 ----------AUGUUC-----GACCUACCCAGCGGAAAUUGUGAUGGAGCUGCAAAUUGGUCGUGUUGCAGGUGGCCUGCGACAUGGUGCCGAUGAUGCAGGGCAGCUUGGUGGCCGGCC ----------....((-----(.((.(((.(((.............(..(((((.(((((((((((((((((...)))))))))))..))))))..)))))..).))).))))).))).. ( -40.24) >DroAna_CAF1 142298 115 - 1 ---UUUUUUUU--CAAGUGUUCUCUUACCCAUCGGAAGUUGUGGUGCAGUUGCAGAUUGGUGGUGUUGCAGGUGGCCUGCGACAUGGUGCCAAUGAUGCAGGGCAGCUUGGUAGCUGGGC ---.......(--(((((((((.((((((((.(....).)).)))).)).((((.(((((((((((((((((...))))))))))..)))))))..))))))))).)))))......... ( -40.90) >consensus ___UU__U______UA_____AACUUACCCAUCGGAAAUUGUGAUGCAGCUGCAGAUUGGUCGUGUUGCAGGUGGCCUGCGACAUGGUGCCAAUGAUGCAGGGCAGCUUGGUGGCUGGCC ............................((..............(((..(((((.(((((((((((((((((...)))))))))))..))))))..))))).)))(((....))).)).. (-25.43 = -26.18 + 0.76)

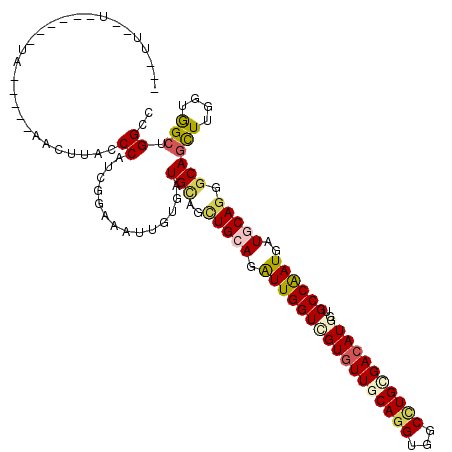
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:47 2006