
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,950,624 – 19,950,756 |
| Length | 132 |
| Max. P | 0.845744 |

| Location | 19,950,624 – 19,950,738 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -46.48 |
| Consensus MFE | -34.53 |
| Energy contribution | -37.24 |
| Covariance contribution | 2.71 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
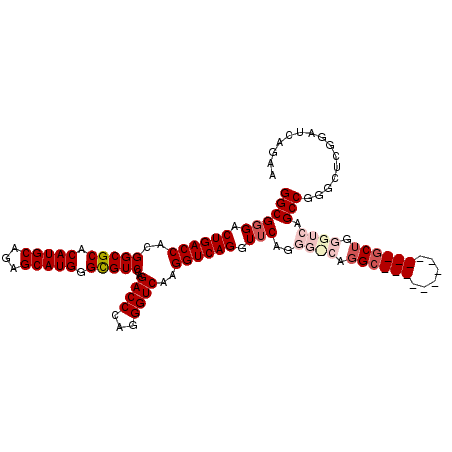
>2L_DroMel_CAF1 19950624 114 + 22407834 GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGCGUCCAGACCCAGGGUCAAGGUCAGGUUCAGGGUCAGGCGCUGGGUCAGGCGCUGGGUCAGCCGGGUUCGGAUCAGAA ..(.((.((((((.((((((.(((((...)))))((((((..(((((.(..((.......))..).))))).)))))).......)))))))))))))).)............. ( -52.70) >DroSec_CAF1 133624 102 + 1 GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGCGUCCAGACCCAGGGUCAAGGUCAGGUUCAAGGCCAGGC------------GCUGGGUCAGCCGGGCUCGGAUCAGAA .....((((((((.((((((.(((((...))))).)).....(((((((.(((..((((........)))).)))------------.))))))).)))))).)))).)).... ( -47.80) >DroSim_CAF1 136133 102 + 1 GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGCGUCCAGACCCAGGGUCAAGGUCAGGUUCAGGGCCAGGC------------GCUGGGUCAGCCGGGCUCGGAUCAAAA .....((((((((.((((((.(((((...))))).)).....(((((((.(((..((((........)))).)))------------.))))))).)))))).)))).)).... ( -47.70) >DroEre_CAF1 132046 90 + 1 GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGCGUCCAGACCCAGGGUCAAGGUCAGCUGCAG------------------------GGUCAGCCGGGCUCAGAGCAGAA .((((..((((((..(((((.(((((...))))).)))))..((((...))))..)))))))))).(------------------------((((.....)))))......... ( -38.10) >DroYak_CAF1 132607 96 + 1 GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGUGUCCAGACCCAGGGUCAAGGUCAGCUUCAG------GC------------CCUGGGUCAGCCGGCCUCAGACCAGAA (((.((.((......(((((.(((((...))))).)))))..(((((((((((.(((....)))..)------))------------)))))))))))).)))........... ( -46.10) >consensus GGCGGGACUGACCACGGCGCACAUGCAGAGCAUGGGCGUCCAGACCCAGGGUCAAGGUCAGGUUCAGGG_CAGGC____________GCUGGGUCAGCCGGGCUCGGAUCAGAA ((((((.((((((..(((((.(((((...))))).)))))..((((...))))..)))))).)))..((((.((((((......)))))).)))).)))............... (-34.53 = -37.24 + 2.71)

| Location | 19,950,664 – 19,950,756 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19950664 92 + 22407834 CAGACCCAGGGUCAAGGUCAGGUUCAGGGUCAGGCGCUGGGUCAGGCGCUGGGUCAGCCGGGUUCGGAUCAGAAGCCACCGAACUCGGGUUC ..(((((((.(((.......((..(((.(.....).)))..)).))).)))))))..((((((((((...........)))))))))).... ( -41.30) >DroSec_CAF1 133664 80 + 1 CAGACCCAGGGUCAAGGUCAGGUUCAAGGCCAGGC------------GCUGGGUCAGCCGGGCUCGGAUCAGAAGCCACCGAACUCGGGUUC ..(((((((.(((..((((........)))).)))------------.)))))))..(((((.((((...........)))).))))).... ( -36.10) >DroSim_CAF1 136173 80 + 1 CAGACCCAGGGUCAAGGUCAGGUUCAGGGCCAGGC------------GCUGGGUCAGCCGGGCUCGGAUCAAAAGCCACCGAACUCGGGUUC ..(((((((.(((..((((........)))).)))------------.)))))))..(((((.((((...........)))).))))).... ( -36.00) >DroYak_CAF1 132647 74 + 1 CAGACCCAGGGUCAAGGUCAGCUUCAG------GC------------CCUGGGUCAGCCGGCCUCAGACCAGAAGCCACCGACCUCGGGCUC ..(((((((((((.(((....)))..)------))------------))))))))(((((((.((......)).)))..((....)))))). ( -36.70) >consensus CAGACCCAGGGUCAAGGUCAGGUUCAGGGCCAGGC____________GCUGGGUCAGCCGGGCUCGGAUCAGAAGCCACCGAACUCGGGUUC ..(((((((.......................................)))))))..(((((.((((...........)))).))))).... (-20.81 = -21.30 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:45 2006