| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,927,771 – 19,927,877 |
| Length | 106 |
| Max. P | 0.899406 |

| Location | 19,927,771 – 19,927,877 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.41 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
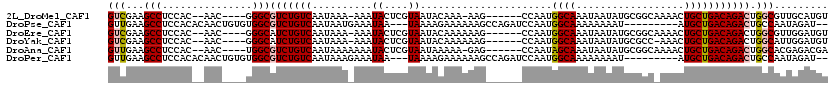
>2L_DroMel_CAF1 19927771 106 - 22407834 GUCGAAGCCUCCAC--AAC----GGGCGUCUGUCAAUAAA-AAAUACUCGUAAUACAAA-AAG------CCAAUGGCAAAUAAUAUGCGGCAAAACUGCUGACAGACUGGCGUUGCAUGU ((((..((((....--...----))))((((((((.....-..................-..(------((...))).........((((.....))))))))))))))))......... ( -27.10) >DroPse_CAF1 176548 106 - 1 GUUGAAGCCUCCACACAACUGUGUGGCGUCUGUCAAUAAUGAAAUAA---UAAAAGAAAAAAGCCAGAUCCAAUGGCAAAAAAAAU---------AUGCUGACAGACUGCCAAUAGAU-- ((((..((..((((((....)))))).((((((((...((......)---)...........((((.......)))).........---------....)))))))).))))))....-- ( -26.30) >DroEre_CAF1 109291 107 - 1 GUCGAAGCCUCCAC--AAC----GGGCAUCUGUCAAUAAA-AAAUACUCGUAAUACAAAAAAG------CCAAUGGCAAAUAAUAUGCGGCAAAACUGCUGACAGACUGGCGUUGGAUGU (((.((((((((..--...----)))..(((((((.....-.....................(------((...))).........((((.....)))))))))))..))).)).))).. ( -24.20) >DroYak_CAF1 109815 106 - 1 GUCGAAGCCUCCAC--AAC----GGGCAUCUGUCAAUAAA-AAAUACUCGUAAUACAAAAAAG------CCAAUGGCAAAUAAUAUGCGCC-AAACUGCUGACAGACUGGCAUUGGAUGU (((.((((((((..--...----)))..((((((......-........(.....).....((------(...((((...........)))-)....)))))))))..))).)).))).. ( -21.50) >DroAna_CAF1 118279 107 - 1 GUUGAAGCCUCCAC--AAC----UGGCGUCUGUCAAUAAAAAAAUACUCGUAAUAAAAA-GAG------CCAAUAGCAAAUAAUAUGCGGCAAAACUGCUGACAGACUGGCACGAGACGA (((...(((.(((.--...----))).((((((((...........(((..........-)))------.................((((.....)))))))))))).)))....))).. ( -24.20) >DroPer_CAF1 194720 106 - 1 GUUGAAGCCUCCACACAACUGUGUGGCGUCUGUCAAUAAAGAAAUAA---UAAAAGAAAAAAGCCAGAUCCAAUGGCAAAAAAAAU---------AUGCUGACAGACUGCCAAUAGAU-- ((((..((..((((((....)))))).((((((((....(....)..---............((((.......)))).........---------....)))))))).))))))....-- ( -26.70) >consensus GUCGAAGCCUCCAC__AAC____GGGCGUCUGUCAAUAAA_AAAUACUCGUAAUACAAAAAAG______CCAAUGGCAAAUAAUAUGCGGC_AAACUGCUGACAGACUGGCAUUAGAUGU (((...(((...............)))(((((((..........((....))......................((((..................))))))))))).)))......... (-13.14 = -13.41 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:40 2006