| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
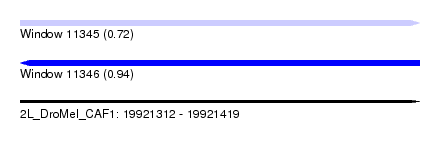
| Location | 19,921,312 – 19,921,419 |
| Length | 107 |
| Max. P | 0.938342 |

| Location | 19,921,312 – 19,921,419 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.17 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
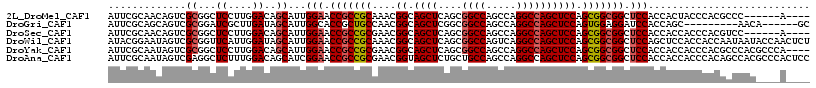
>2L_DroMel_CAF1 19921312 107 + 22407834 AUUCGCAACAGUCGCGGCUCCUUGGACAGCAUUGGAACCGCCGCAAACGGCAGCUCAGCGGCCAGCCAGGCCAGCUCCAGCGGCGGCUCCACCACUACCCACGCCC------A---- ....((.......))(((....(((..((...((((.(((((((....((.((((....((((.....)))))))))).))))))).))))...))..))).))).------.---- ( -40.70) >DroGri_CAF1 136787 102 + 1 AUUCGCAGCAGUCGCGGAUCGCUUGAUAGCAUUGGCACCGCUGCCAACGGCAGCUCGGCGGCCAGCCAGGCCAGCUCCAGUGGAGGAUCCACCAGC---------AACA------GC ....((.((....))((((((((....))).(((((...((((((...))))))..(((.....)))..)))))((((...)))))))))....))---------....------.. ( -40.30) >DroSec_CAF1 97362 107 + 1 AUUCGCAACAGUCGCGGCUCCUUGGACAGCAUUGGAACCGCCGCGAACGGCAGCUCAGCGGCCAGCCAGGCCAGCUCCAGCGGCGGCUCCACCACCACCCACGUCC------A---- ..((((.......)))).....(((((.....((((.(((((((....((.((((....((((.....)))))))))).))))))).))))...........))))------)---- ( -40.09) >DroWil_CAF1 170252 117 + 1 AUACGGAAUAGUCGCGGUUCAUUGGAUAGCAUUGGAACCGCCGCAAACGGCAGCUCAGCGGCCAGUCAGGCCAGCUCCAGCGGCGGCUCCAGCUCCACCACCAAUAAUACCAACUCU ...............(((..(((((..(((..((((.(((((((....((.((((....((((.....)))))))))).))))))).)))))))......)))))...)))...... ( -42.00) >DroYak_CAF1 103349 113 + 1 AUUCGCAAUAGUCGCGGCUCCUUGGACAGCAUUGGAACCGCCGCGAACGGCAGCUCAGCGGCCAGCCAGGCCAGCUCCAGCGGCGGCUCCACCACCACCCACGCCCACGCCCA---- ....((.....((((((((((.((.....))..)))...)))))))..(((.(((.(((((((.....)))).)))..)))((.((.....)).))......)))...))...---- ( -40.70) >DroAna_CAF1 111571 117 + 1 AUUCGCAAUAGUCGAGGCUCUUUGGACAGCAUCGGAACCGCCGCGAACGGUAGCUCUGCUGCCAGCCAGGCCAGCUCCAGCGGCGGCUCCACCACCACCCACAGCCACGCCCACUCC ..((((....).)))((((...(((........(((.(((((((....((((((...))))))(((.......)))...))))))).)))........))).))))........... ( -38.99) >consensus AUUCGCAACAGUCGCGGCUCCUUGGACAGCAUUGGAACCGCCGCGAACGGCAGCUCAGCGGCCAGCCAGGCCAGCUCCAGCGGCGGCUCCACCACCACCCACGCCCAC____A____ .............((...((....))..))...(((.(((((((....((.((((....((((.....)))))))))).))))))).)))........................... (-31.78 = -32.17 + 0.39)


| Location | 19,921,312 – 19,921,419 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -56.05 |
| Consensus MFE | -39.52 |
| Energy contribution | -41.97 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19921312 107 - 22407834 ----U------GGGCGUGGGUAGUGGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUUGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUGUUGCGAAU ----.------.(((.(((..(((..(((((((((((((((((((((((.....))))....)))).)))...))))))))))))..)))..)))....)))((((...)))).... ( -57.50) >DroGri_CAF1 136787 102 - 1 GC------UGUU---------GCUGGUGGAUCCUCCACUGGAGCUGGCCUGGCUGGCCGCCGAGCUGCCGUUGGCAGCGGUGCCAAUGCUAUCAAGCGAUCCGCGACUGCUGCGAAU ((------.(((---------((.(((((.....)))))((((((....((((((((.(((..((((((...)))))))))))))..))))...)))..)))))))).))....... ( -43.80) >DroSec_CAF1 97362 107 - 1 ----U------GGACGUGGGUGGUGGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUGUUGCGAAU ----.------((.(.((((..((..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..))..)))).)...))((((...)))).... ( -56.10) >DroWil_CAF1 170252 117 - 1 AGAGUUGGUAUUAUUGGUGGUGGAGCUGGAGCCGCCGCUGGAGCUGGCCUGACUGGCCGCUGAGCUGCCGUUUGCGGCGGUUCCAAUGCUAUCCAAUGAACCGCGACUAUUCCGUAU ..((((((..((((((((((((....(((((((((((((((((((((((.....))))....)))).)))...)))))))))))).))))).)))))))..).)))))......... ( -54.00) >DroYak_CAF1 103349 113 - 1 ----UGGGCGUGGGCGUGGGUGGUGGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUAUUGCGAAU ----(((.(((((.(.((((..((..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..))..))))....)))))).)))........ ( -61.00) >DroAna_CAF1 111571 117 - 1 GGAGUGGGCGUGGCUGUGGGUGGUGGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCAGCAGAGCUACCGUUCGCGGCGGUUCCGAUGCUGUCCAAAGAGCCUCGACUAUUGCGAAU (.(((((.((.((((.((((..((..((((((((((((....((((.((.....)))))).((((....))))))))))))))))..))..))))...)))).)).))))).).... ( -63.90) >consensus ____U____GUGGGCGUGGGUGGUGGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUAUUGCGAAU ................(((((((((.(((((((((((((((((((((((.....))))....)))).)))...)))))))))))).)))))))))......((((.....))))... (-39.52 = -41.97 + 2.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:38 2006