| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,904,922 – 19,905,016 |
| Length | 94 |
| Max. P | 0.740000 |

| Location | 19,904,922 – 19,905,016 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
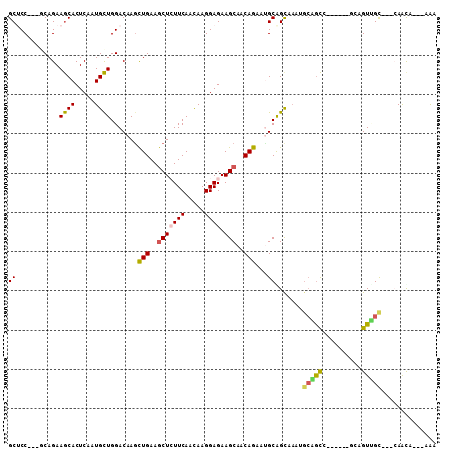
| Mean pairwise identity | 76.27 |
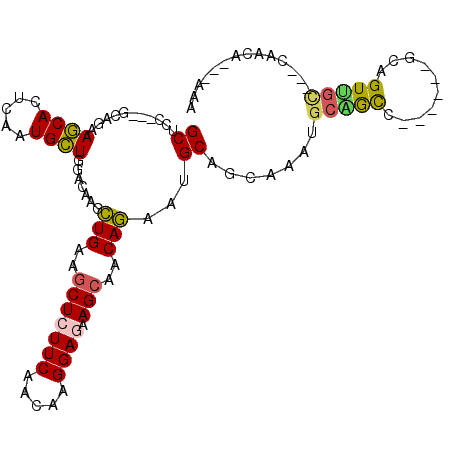
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
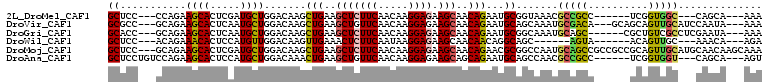
>2L_DroMel_CAF1 19904922 94 + 22407834 GCUCC---CCAGAAGCACUCGAUGCUGGACAAGCUGAAGCUCUUCAACAAGGAGAAGCAACAGAAUGCGGUAAACGCCGCC------UCGGUGGC---CAGCA---AAA (((..---.....)))......((((((....(((((.(((((((.....)))).)))........(((((....))))).------)))))..)---)))))---... ( -31.70) >DroVir_CAF1 133934 100 + 1 GCGCC---GCAGAAGCACUCAAUGCUGGACAAGCUGAAGCUGUUCAACAAGGAGAAGCAACAGAAUGCAGCAAAUGCGACA---GCAGCAGUUGCAUCCAAUA---AAA ((...---((....)).(((..((.((((((.((....)))))))).))..)))..))....(.(((((((...(((....---)))...))))))).)....---... ( -29.90) >DroGri_CAF1 120876 97 + 1 GCACC---GCAGAAGCACUCAAUGCUGGACAAGCUGAAGCUCUUCAACAAGGAGAAGCAACAGAAUGCGGCAAAUGCAGC------CGCUGUCGCCUCGAAUA---AAA .....---((...((((.....)))).((((..(((..(((((((.....)))).)))..)))...(((((.......))------)))))))))........---... ( -27.90) >DroWil_CAF1 148625 88 + 1 GCUCC---ACAGAAACACUCCAUGUUGGACAAGUUGAAACUCUUCAAUAAGGAGAAGCAACAACAGGCAGC------AGUA------ACAGUUGC---AAACA---AGA .....---.........((((.(((((((..(((....))).))))))).))))..(((((.....((...------.)).------...)))))---.....---... ( -18.20) >DroMoj_CAF1 218808 106 + 1 GCUCC---GCAGAAGCACUCGAUGCUGGACAAGCUGAAGCUCUUCAACAAGGAGAAGCAACAGAACGCGGCCAAUGCAGCCGCCGCCGCAGUUGCAUGCAACAAGCAAA (((((---((...((((.....)))).......(((..(((((((.....)))).)))..)))...))))...(((((((.((....)).)))))))......)))... ( -32.60) >DroAna_CAF1 96454 97 + 1 GCUCCUGUCCAGAAGCACUCCAUGCUGGACAAACUGAAGCUGUUCAACAAGGAGAAGCAGCAGAAUGCAGCCAACGCCGCC------UCGGUGGU---CAGCA---AGU (((.((....)).))).((((.((.((((((.........)))))).)).))))..(((......))).((....(((((.------...)))))---..)).---... ( -28.10) >consensus GCUCC___GCAGAAGCACUCAAUGCUGGACAAGCUGAAGCUCUUCAACAAGGAGAAGCAACAGAAUGCAGCAAAUGCAGCC______GCAGUUGC___CAACA___AAA ((...........((((.....)))).......(((..(((((((.....)))).)))..)))...)).......(((((..........))))).............. (-14.15 = -13.82 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:35 2006