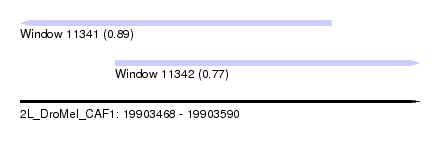
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,903,468 – 19,903,590 |
| Length | 122 |
| Max. P | 0.887864 |

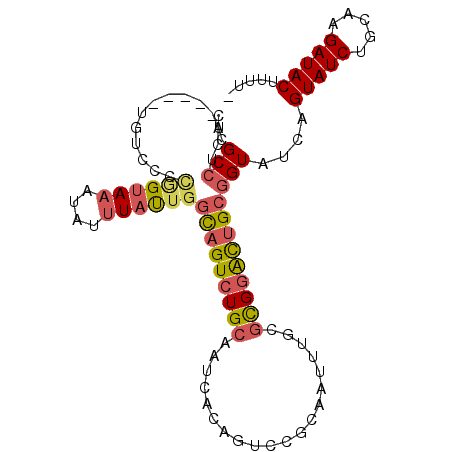
| Location | 19,903,468 – 19,903,563 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19903468 95 - 22407834 CACGCCUCU-----UGUCCCCGGUAAAUAUUUAUUGGCAGUCUGCAAUCACAGUCCGCAAUUUGCGCGGACUGCGGUAUCAGUAUCUGCAAGAUACUUUU- ......(((-----((...(((((((....)))))))(((.(((..(((.((((((((.......)))))))).)))..)))...)))))))).......- ( -30.10) >DroVir_CAF1 132148 93 - 1 CACGCGGCUCUCUCAGGCCGUAGAACACAUUUACAGCUUGUCUGG--------GGCGUGAUUUGCAUGAGUUGCGGUCACAGUAUCUGUCAGAUACAUUUU ......(((((..(((((.(((((.....))))).)))))...))--------)))((((((.(((.....))))))))).(((((.....)))))..... ( -30.60) >DroSec_CAF1 79923 95 - 1 CACGCCUCU-----UGUCCCCGGUAAAUAUUUAUUGGCAGUCUGCAAUCACAGUCCGCAAUUUGCGCGGACUGCGGUAUCAGUAUCUGCAAGAUACUUUU- ......(((-----((...(((((((....)))))))(((.(((..(((.((((((((.......)))))))).)))..)))...)))))))).......- ( -30.10) >DroSim_CAF1 91140 95 - 1 CACGCCUCU-----UGUCCCCGGUAAAUAUUUAUUGGCAGUCUGCAAUCACAGUCCGCAAUUUGCGCGGACUGCGGUAUCAGUAUCUGCAAGAUACUUUU- ......(((-----((...(((((((....)))))))(((.(((..(((.((((((((.......)))))))).)))..)))...)))))))).......- ( -30.10) >DroEre_CAF1 86701 89 - 1 CACGCCUCU-----GGUCCCAAGUAAAUAUUUAUUGGCAGUCUGCAAUCACAG------AUUUGCGCGGACUGCGGUAUCAGUAUCUGUAAGAUACUUUU- ........(-----(((.((.(((((....))))).(((((((((((((...)------)))...))))))))))).))))(((((.....)))))....- ( -24.70) >DroYak_CAF1 85016 89 - 1 CACGCCUCU-----GGUUCCCGGUAAAUAUUUAUUGGCAGUCUGCAAUCACAG------AUUUGCGUGGACUGCGGUAUCAGUAUCUGUAAGAUACUUUU- ........(-----(((.((((((((....))))))(((((((((((......------..))))..))))))))).))))(((((.....)))))....- ( -25.00) >consensus CACGCCUCU_____UGUCCCCGGUAAAUAUUUAUUGGCAGUCUGCAAUCACAGUCCGCAAUUUGCGCGGACUGCGGUAUCAGUAUCUGCAAGAUACUUUU_ ...(((..............((((((....))))))(((((((((....................))))))))))))....(((((.....)))))..... (-16.88 = -17.38 + 0.50)


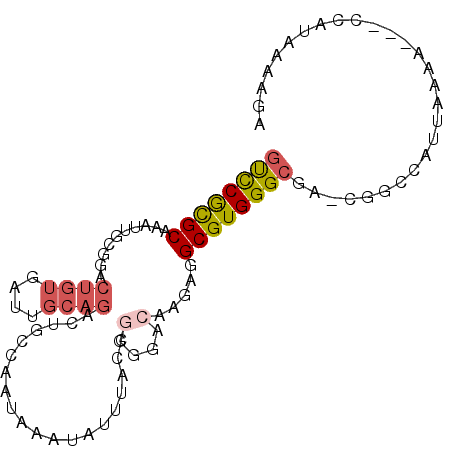
| Location | 19,903,497 – 19,903,590 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.22 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -14.59 |
| Energy contribution | -15.12 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19903497 93 + 22407834 GUCCGCGCAAAUUGCGGACUGUGAUUGCAGACUGCCAAUAAAUAUUUACCGGGGACAAGAGGCGUGGGCAA-CGACCAUUAAAA---UCAUAAAAGA ((((((((.....((((.((((....)))).))))...............(....).....))))))))..-............---.......... ( -26.30) >DroGri_CAF1 119107 84 + 1 ACUCAUGCAAAUCACGCC--------UCAGACAAGCUGUAACUGCGCU---UGGCCU-ACCGCGUGGGCGGUUUGCCAUAAAA-AAGCCAUAAAGAA ......(((((((.....--------.....(((((.((....)))))---))((((-((...))))))))))))).......-............. ( -22.70) >DroSec_CAF1 79952 93 + 1 GUCCGCGCAAAUUGCGGACUGUGAUUGCAGACUGCCAAUAAAUAUUUACCGGGGACAAGAGGCGUGGGCGA-CGGCCAUUAAAA---UCAUAAAAGA ((((((((.....((((.((((....)))).))))...............(....).....))))))))..-............---.......... ( -26.30) >DroSim_CAF1 91169 93 + 1 GUCCGCGCAAAUUGCGGACUGUGAUUGCAGACUGCCAAUAAAUAUUUACCGGGGACAAGAGGCGUGGGCGA-CGGCCAUUAAAA---UCAUAAAAGA ((((((((.....((((.((((....)))).))))...............(....).....))))))))..-............---.......... ( -26.30) >DroEre_CAF1 86730 87 + 1 GUCCGCGCAAAU------CUGUGAUUGCAGACUGCCAAUAAAUAUUUACUUGGGACCAGAGGCGUGGGCGA-AGGCCAUUAAAA---CCAUAAAAGA ((((((((...(------((((....)))))((((((((((....))).))))...)))..))))))))..-............---.......... ( -23.40) >DroYak_CAF1 85045 87 + 1 GUCCACGCAAAU------CUGUGAUUGCAGACUGCCAAUAAAUAUUUACCGGGAACCAGAGGCGUGGGCGA-CGGCCAUUAAAA---CCAUAAAAGA ((((((((...(------((((....)))))(((((..(((....)))...))...)))..))))))))..-............---.......... ( -21.60) >consensus GUCCGCGCAAAUUGCGGACUGUGAUUGCAGACUGCCAAUAAAUAUUUACCGGGGACAAGAGGCGUGGGCGA_CGGCCAUUAAAA___CCAUAAAAGA ((((((((..........((((....))))....................(....).....))))))))............................ (-14.59 = -15.12 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:34 2006