| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,867,376 – 19,867,469 |
| Length | 93 |
| Max. P | 0.993323 |

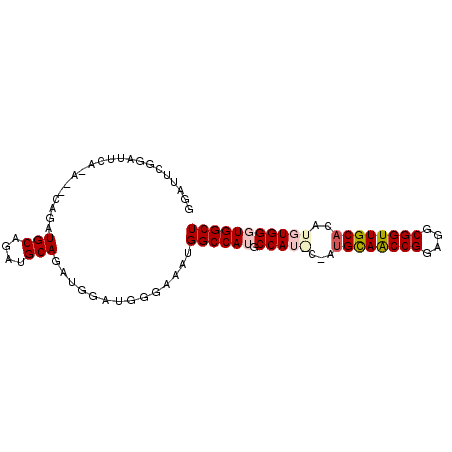
| Location | 19,867,376 – 19,867,469 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -18.11 |
| Energy contribution | -19.16 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
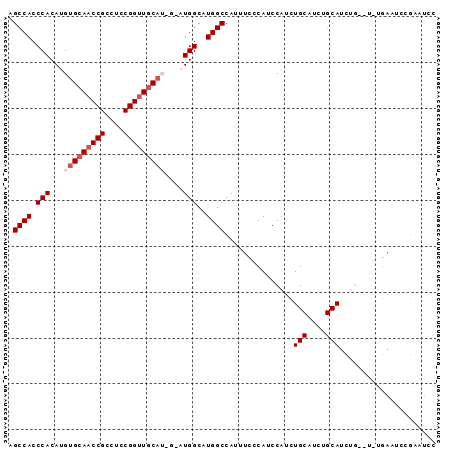
>2L_DroMel_CAF1 19867376 93 + 22407834 AGCCACCCACAUGAGCAACCGCCUCCGGUUACAUUGCAUGGCAUGGCCAUUUCCCAUCCAUCUGCAUCUGCAUCUGACUCUGAAUCCGAAUCC .........((.(((((((((....)))))....(((((((.((((.......)))))))..))))...)).))))................. ( -17.00) >DroSec_CAF1 44121 87 + 1 AGCCACCCACAUGUGCAACCGCCUCCGGUUGCAUGGUAUGGCAUGGCCAGUUCACAUCCAUCUGCAUCUGCAUC------UGAAUCCGAAUCC .((((.(((..((((((((((....))))))))))...)))..))))..((((..((.((..(((....)))..------)).))..)))).. ( -26.00) >DroSim_CAF1 53737 81 + 1 AGCCACCCACAUGUGCAACCGCCUCCGGCUGCAUGGCAUGGCAUGGCCAUUUCACAUCCAUCUG------CAUC------CGAAUCCGAAUCC .((((.(((..((((((.(((....))).))))))...)))..)))).................------....------............. ( -18.00) >DroEre_CAF1 51380 87 + 1 AGCCACCCACAUGUGCAACCGCCUCCGGUUGCA-----UGGCCUGGCCACUUUCCAUCCAUCUGCAUCUGCAUCUGCAUCUGACUCUGAG-UC .((((.(((....((((((((....))))))))-----)))..)))).((((......((..((((........))))..)).....)))-). ( -23.40) >DroYak_CAF1 49588 76 + 1 AGCCACCCACAUGUGCAACCGCCUCCGGUUGCA-----UGGCUUGGCCAUUUCCCAUCCAUCUGCAUCUGCAUCUGAGU------------UC .((((.(((....((((((((....))))))))-----)))..))))...........((..(((....)))..))...------------.. ( -22.10) >consensus AGCCACCCACAUGUGCAACCGCCUCCGGUUGCAU_G_AUGGCAUGGCCAUUUCCCAUCCAUCUGCAUCUGCAUCUG__U_UGAAUCCGAAUCC .((((.(((...(((((((((....)))))))))....)))..))))...............(((....)))..................... (-18.11 = -19.16 + 1.05)

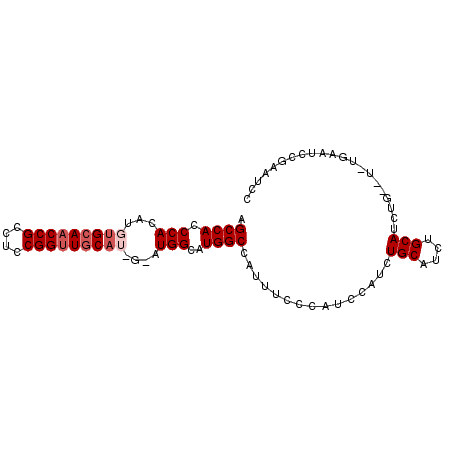
| Location | 19,867,376 – 19,867,469 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.96 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19867376 93 - 22407834 GGAUUCGGAUUCAGAGUCAGAUGCAGAUGCAGAUGGAUGGGAAAUGGCCAUGCCAUGCAAUGUAACCGGAGGCGGUUGCUCAUGUGGGUGGCU .........(((....(((..(((....)))..)))....)))..((((((.(((((....(((((((....))))))).)))).).)))))) ( -31.40) >DroSec_CAF1 44121 87 - 1 GGAUUCGGAUUCA------GAUGCAGAUGCAGAUGGAUGUGAACUGGCCAUGCCAUACCAUGCAACCGGAGGCGGUUGCACAUGUGGGUGGCU ((.((((.(((((------..(((....)))..))))).))))))((((((.(((((...((((((((....))))))))..))))))))))) ( -39.20) >DroSim_CAF1 53737 81 - 1 GGAUUCGGAUUCG------GAUG------CAGAUGGAUGUGAAAUGGCCAUGCCAUGCCAUGCAGCCGGAGGCGGUUGCACAUGUGGGUGGCU ...((((.(((((------....------....))))).))))..((((((.(((((...((((((((....)))))))))))).).)))))) ( -32.60) >DroEre_CAF1 51380 87 - 1 GA-CUCAGAGUCAGAUGCAGAUGCAGAUGCAGAUGGAUGGAAAGUGGCCAGGCCA-----UGCAACCGGAGGCGGUUGCACAUGUGGGUGGCU ((-(.....)))..((.((..(((....)))..)).))....(((.(((..((..-----((((((((....))))))))...)).))).))) ( -29.30) >DroYak_CAF1 49588 76 - 1 GA------------ACUCAGAUGCAGAUGCAGAUGGAUGGGAAAUGGCCAAGCCA-----UGCAACCGGAGGCGGUUGCACAUGUGGGUGGCU ..------------.((((..(((....)))......))))....(((((..(((-----((((((((....))))))))....))).))))) ( -27.50) >consensus GGAUUCGGAUUCA_A__CAGAUGCAGAUGCAGAUGGAUGGGAAAUGGCCAUGCCAU_C_AUGCAACCGGAGGCGGUUGCACAUGUGGGUGGCU .....................(((....)))..............((((((.(((((...((((((((....))))))))..))))))))))) (-22.91 = -23.96 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:29 2006