
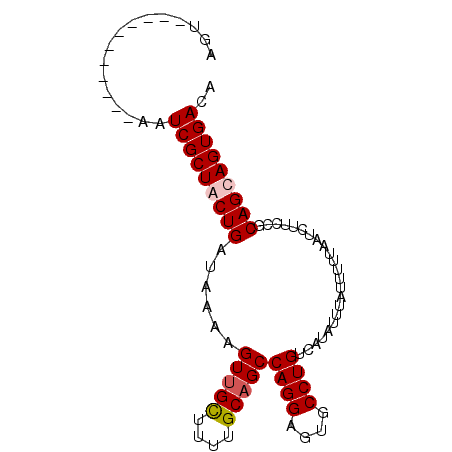
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,836,856 – 19,836,947 |
| Length | 91 |
| Max. P | 0.919886 |

| Location | 19,836,856 – 19,836,947 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19836856 91 + 22407834 AGUAAAGUAAAUGUAAUCGCUGCUGAUAAAAGUUGCUUUUGCAGCCAGGAGUGCCUGUCAUAUUUAUUUUUAAUCUUCCGCAGCAGUGACA ................(((((((((......(((((....)))))((((....)))).......................))))))))).. ( -25.20) >DroSim_CAF1 21299 80 + 1 GGC-----------AAUCGCUGCUGAUAAAAGUUGCUUUUGCAGCCAGGAGUGCCUGUCAUAUUUAUUUUUAAUCUUCCGCAGCAGUGACA ...-----------..(((((((((......(((((....)))))((((....)))).......................))))))))).. ( -25.20) >DroEre_CAF1 20917 80 + 1 AGU-----------AAUCGCUACUGAUAAAAGUUGUUCUUGCUGCCAGGAGUGCCUGUCAUAUUUAUUUUUAAUCUUCCGCAGCAGUGACA (((-----------(.....)))).........((((.(((((((((((....))))......(((....)))......))))))).)))) ( -16.60) >DroYak_CAF1 18122 80 + 1 AGU-----------AAUCGCUACUGAUAAAAGUUGUUUUUGCAGCCAGGAGUGCCUGUCAUAUUUAUUUUUAAUCUUCCGCAGCAGUGACA ...-----------..(((((.(((......(((((....)))))((((....)))).......................))).))))).. ( -16.50) >consensus AGU___________AAUCGCUACUGAUAAAAGUUGCUUUUGCAGCCAGGAGUGCCUGUCAUAUUUAUUUUUAAUCUUCCGCAGCAGUGACA ................(((((((((......(((((....)))))((((....)))).......................))))))))).. (-20.23 = -20.72 + 0.50)


| Location | 19,836,856 – 19,836,947 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -18.70 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19836856 91 - 22407834 UGUCACUGCUGCGGAAGAUUAAAAAUAAAUAUGACAGGCACUCCUGGCUGCAAAAGCAACUUUUAUCAGCAGCGAUUACAUUUACUUUACU .(((.((((((.(((((.................((((....))))(((.....)))..)))))..)))))).)))............... ( -21.80) >DroSim_CAF1 21299 80 - 1 UGUCACUGCUGCGGAAGAUUAAAAAUAAAUAUGACAGGCACUCCUGGCUGCAAAAGCAACUUUUAUCAGCAGCGAUU-----------GCC .(((.((((((.(((((.................((((....))))(((.....)))..)))))..)))))).))).-----------... ( -21.80) >DroEre_CAF1 20917 80 - 1 UGUCACUGCUGCGGAAGAUUAAAAAUAAAUAUGACAGGCACUCCUGGCAGCAAGAACAACUUUUAUCAGUAGCGAUU-----------ACU .(((.(((((((....)..............((.((((....)))).)).................)))))).))).-----------... ( -15.30) >DroYak_CAF1 18122 80 - 1 UGUCACUGCUGCGGAAGAUUAAAAAUAAAUAUGACAGGCACUCCUGGCUGCAAAAACAACUUUUAUCAGUAGCGAUU-----------ACU .(((.(((((((....)..............((.(((.((....)).)))))..............)))))).))).-----------... ( -15.90) >consensus UGUCACUGCUGCGGAAGAUUAAAAAUAAAUAUGACAGGCACUCCUGGCUGCAAAAACAACUUUUAUCAGCAGCGAUU___________ACU .(((.((((((.(((((.................((((....))))(((.....)))..)))))..)))))).)))............... (-17.20 = -17.70 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:20 2006