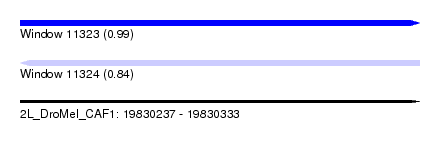
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,830,237 – 19,830,333 |
| Length | 96 |
| Max. P | 0.988690 |

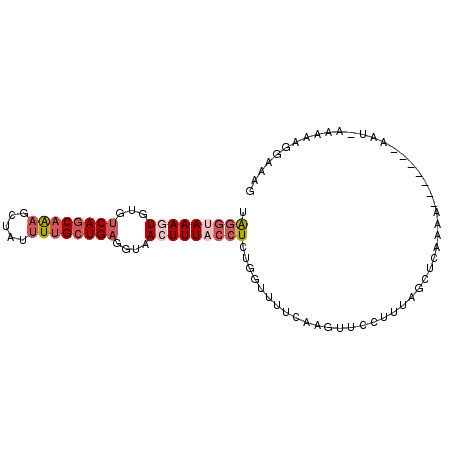
| Location | 19,830,237 – 19,830,333 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -11.45 |
| Energy contribution | -13.28 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19830237 96 + 22407834 CUUUCCUUUUU-AUUUUAUUUUUUUUGAGCUAAAGGAACUUGAAAACCAGAGGAAAAGUUACCUCAGCAAAAUAGCUUUGCUGACACACUUUACCCA ...((((((..-.(((((..((((((......))))))..)))))...))))))(((((....((((((((.....))))))))...)))))..... ( -21.00) >DroSec_CAF1 7113 89 + 1 CUUUCCUUUUU-AUU-------UUUUGAGCUGAGGGAACUUGAAAGCCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACACACUUUACCUA ...........-...-------.((((.((((((....)))...)))))))((((((((....((((((((.....))))))))...)))))))).. ( -26.00) >DroSim_CAF1 14708 89 + 1 CUUUCCUUUUU-AUU-------UUUGGAGCUGAGGGAACUUAAAAGCCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACACACUUUACCCA ...........-...-------(((((...((((....))))....)))))((((((((....((((((((.....))))))))...)))))))).. ( -26.80) >DroEre_CAF1 14366 83 + 1 CU-----UGU--UUU-------UUUGGGACUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACACAAUUUACCUA (.-----.((--(((-------((((....)))))))))..)........(((((((......((((((((.....)))))))).....))))))). ( -22.00) >DroYak_CAF1 11607 83 + 1 CU-----UUU--UUU-------UUCGGAAGUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGGUUUGCUGACACACUUUACCUA ..-----(((--(((-------(.(....).)))))))............(((((((((....((((((((.....))))))))...))))))))). ( -23.90) >DroAna_CAF1 22234 89 + 1 UUUUCCUUUUCUGUU------UUUUUAAGCCC--GAUUCCUAGAAAGUGGAGAGAAAGUUGCCUCAGCAAAAAUGCGCAGAUGCCACAGUUUAAAUA (((((((((((((..------...........--......))))))).))))))...((.((.((.(((....)))...)).)).)).......... ( -14.11) >consensus CUUUCCUUUUU_AUU_______UUUGGAGCUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACACACUUUACCUA ...................................................((((((((....((((((((.....))))))))...)))))))).. (-11.45 = -13.28 + 1.83)

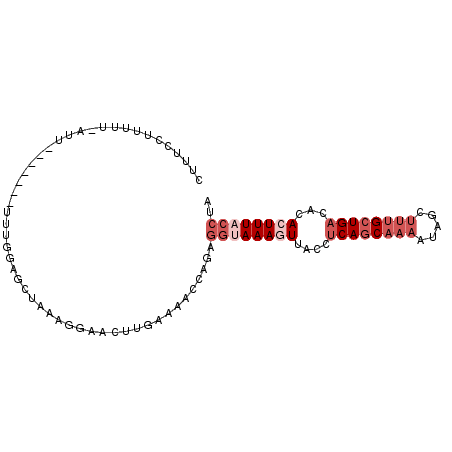
| Location | 19,830,237 – 19,830,333 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -13.19 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19830237 96 - 22407834 UGGGUAAAGUGUGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUUCCUCUGGUUUUCAAGUUCCUUUAGCUCAAAAAAAAUAAAAU-AAAAAGGAAAG .(((.(((((...(((((((((...)))))))))....))))).)))............(((((((...................-..))))))).. ( -19.70) >DroSec_CAF1 7113 89 - 1 UAGGUAAAGUGUGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGCUUUCAAGUUCCCUCAGCUCAAAA-------AAU-AAAAAGGAAAG .(((((((((...(((((((((...)))))))))....)))))))))............((((............-------...-.....)))).. ( -20.95) >DroSim_CAF1 14708 89 - 1 UGGGUAAAGUGUGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGCUUUUAAGUUCCCUCAGCUCCAAA-------AAU-AAAAAGGAAAG .(((((((((...(((((((((...)))))))))....)))))))))...(((.............)))(((...-------...-.....)))... ( -21.12) >DroEre_CAF1 14366 83 - 1 UAGGUAAAUUGUGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUAGUCCCAAA-------AAA--ACA-----AG .((((((((((..(((((((((...)))))))))..))).))))))).(((..((.(((...))).))..)))..-------...--...-----.. ( -19.50) >DroYak_CAF1 11607 83 - 1 UAGGUAAAGUGUGUCAGCAAACCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUACUUCCGAA-------AAA--AAA-----AG .(((((((((...((((((((.....))))))))....)))))))))....(((((((((......))))..)))-------)).--...-----.. ( -23.20) >DroAna_CAF1 22234 89 - 1 UAUUUAAACUGUGGCAUCUGCGCAUUUUUGCUGAGGCAACUUUCUCUCCACUUUCUAGGAAUC--GGGCUUAAAAA------AACAGAAAAGGAAAA ........((((.((....))...(((..((((((....)))....(((........)))...--.)))..)))..------.)))).......... ( -13.30) >consensus UAGGUAAAGUGUGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUAGCUCAAAA_______AAU_AAAAAGGAAAG .(((((((((...((((((((.....))))))))....))))))))).................................................. (-13.19 = -14.50 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:19 2006