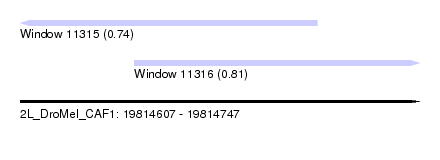
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,814,607 – 19,814,747 |
| Length | 140 |
| Max. P | 0.808228 |

| Location | 19,814,607 – 19,814,711 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -6.39 |
| Energy contribution | -9.60 |
| Covariance contribution | 3.21 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19814607 104 - 22407834 ACAAGGUAAAAGCCAAACUGAAA--GUUGAA--------------UCAGCUUUUACCUAUGCGUUUCAUCUUAGCACCAUGAGUCAUUUUAAAAUAAAAGGAAAAGGGUAGCAGAAUAUU ...((((((((((..(((.....--)))...--------------...)))))))))).((((((.......)))(((.....((.(((((...))))).))....))).)))....... ( -20.00) >DroSec_CAF1 81171 118 - 1 ACAAGGUAAAAGCCAAAAUGAAA--GUUUAAACACUUUUCACAAGUCAGCUUUUACCUAUGCGUUUAAUCUCAGAUCCACGAGUCAUUUUAAAAUAAAAAGAAAAAGCUUGCAUAAUAUU ....(((((((((.....(((((--((......)).))))).......)))))))))(((((.(((((.(((........))).....))))).....(((......))))))))..... ( -19.90) >DroSim_CAF1 83208 118 - 1 ACAAGGUAAGAGCCAAACUGAAA--GUUGAAACACUUUUCAACAGUCAGCUUUUACCUAUGCGUUUAAUCGCAGAUCCACGAGUCAUUUUAAAAUAAAAAUAAAAGGCUUGCAGAAUAUU ...((((((((((.....(((..--(((((((....)))))))..))))))))))))).((((......))))..((..((((((.(((((.........)))))))))))..))..... ( -32.50) >DroEre_CAF1 83670 120 - 1 ACAAGGAAAAAGACAAACUGAAAGAGUUGAAACACUUUUCAGCAGUCAGCUUUUAACUAGGCGUUUAAUCUCAGAUGUACUAGUAGUUGCUAAAUUAAAAUGAAAGGCUUGCAGAAUAUU .((((......(((...(((((.((((......)))))))))..)))(((..(((.((((((((((......)))))).)))))))..)))................))))......... ( -23.20) >DroYak_CAF1 90998 120 - 1 ACAAGGAAAAAGACAAGCCGAAAGAGUUGAAACACUUUUCAACAGUAAGCUUUCAGCUAUGCGUUUAAUCUCUGAUGCGCGAGUAGUUUUAAAAUUAAAAUGUAAGGCUUGCUGAACAUU ....(((((......((((....).))).......)))))..((((((((((((.(((.(((((............)))))))).((((((....))))))).)))))))))))...... ( -27.12) >consensus ACAAGGUAAAAGCCAAACUGAAA__GUUGAAACACUUUUCAACAGUCAGCUUUUACCUAUGCGUUUAAUCUCAGAUCCACGAGUCAUUUUAAAAUAAAAAUAAAAGGCUUGCAGAAUAUU ...(((((((((.....((((....(((((((....)))))))..))))))))))))).............................................................. ( -6.39 = -9.60 + 3.21)

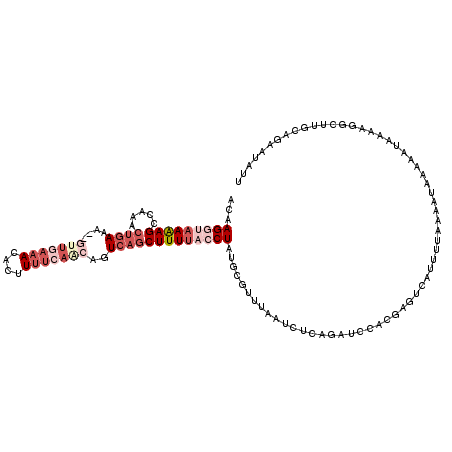
| Location | 19,814,647 – 19,814,747 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -10.46 |
| Energy contribution | -14.14 |
| Covariance contribution | 3.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19814647 100 + 22407834 AUGGUGCUAAGAUGAAACGCAUAGGUAAAAGCUGA--------------UUCAAC--UUUCAGUUUGGCUUUUACCUUGUA----UUAUCUAGUCCCAUUUUCAAUUUCUUUCAUCGUUU ((((..(((.(((((.(((...((((((((((..(--------------......--.......)..))))))))))))).----))))))))..))))..................... ( -28.12) >DroSec_CAF1 81211 118 + 1 GUGGAUCUGAGAUUAAACGCAUAGGUAAAAGCUGACUUGUGAAAAGUGUUUAAAC--UUUCAUUUUGGCUUUUACCUUGUAUUUAUUAUCUAGUCCCAUUUUCAAUUUCUUUCAUCGUUU (.((((..((...((((..((.((((((((((..(...((((((.((......))--)))))).)..))))))))))))..))))...))..)))))....................... ( -33.40) >DroSim_CAF1 83248 114 + 1 GUGGAUCUGCGAUUAAACGCAUAGGUAAAAGCUGACUGUUGAAAAGUGUUUCAAC--UUUCAGUUUGGCUCUUACCUUGUA----UUAUCUAGUCCCAUUUUCAAUUUCUUUCAUCGUUU ((((..(((.(((.....(((.(((((((((((((..(((((((....)))))))--..))))))).....))))))))).----..))))))..))))..................... ( -27.40) >DroEre_CAF1 83710 116 + 1 GUACAUCUGAGAUUAAACGCCUAGUUAAAAGCUGACUGCUGAAAAGUGUUUCAACUCUUUCAGUUUGUCUUUUUCCUUGUA----UUAUCUGCGUCCAUUUUCAAUUUCUUUCAUCGUUU .......(((((....((((.((((...(((..(((.((((((((((......))).)))))))..)))......)))..)----)))...))))....)))))................ ( -20.10) >DroYak_CAF1 91038 116 + 1 GCGCAUCAGAGAUUAAACGCAUAGCUGAAAGCUUACUGUUGAAAAGUGUUUCAACUCUUUCGGCUUGUCUUUUUCCUUGUA----UUAUCUGUGCCCAUUUUCAAUUUCUUUCAUCGUUU (((((.(((.((..(((.(((.(((((((((......(((((((....))))))).)))))))))))).))).)).)))..----.....)))))......................... ( -23.40) >consensus GUGGAUCUGAGAUUAAACGCAUAGGUAAAAGCUGACUGUUGAAAAGUGUUUCAAC__UUUCAGUUUGGCUUUUACCUUGUA____UUAUCUAGUCCCAUUUUCAAUUUCUUUCAUCGUUU ((((..(((.(((.....(((.(((((((((((((..(((((((.............))))))))))))))))))))))).......))))))..))))..................... (-10.46 = -14.14 + 3.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:11 2006