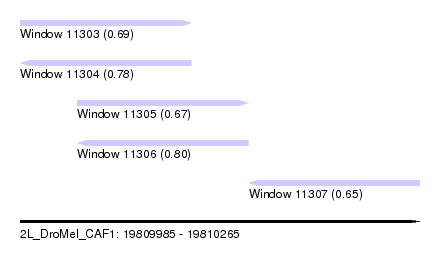
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,809,985 – 19,810,265 |
| Length | 280 |
| Max. P | 0.795868 |

| Location | 19,809,985 – 19,810,105 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -35.74 |
| Energy contribution | -35.63 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
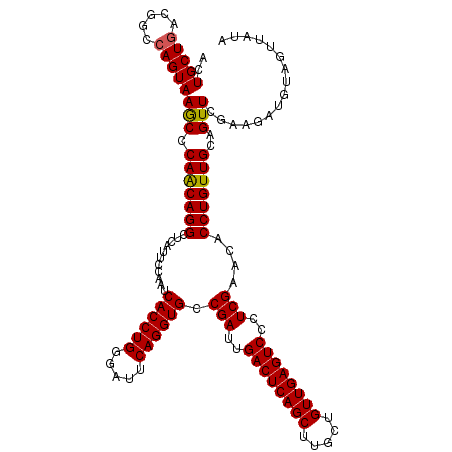
>2L_DroMel_CAF1 19809985 120 + 22407834 AGUGCUAACGCCCAGUAAACCCAGCAGGCUCAUUCCAAUCACCUGGGAUUCAGGUGCCGAUUGACUCAGCUCGCUGUUGAGUCCCUCGAAUACCUGUUGCAGUUCGUAGAUAUAGUUAUA .(.((....)).).(..(((.(((((((...........((((((.....)))))).(((..((((((((.....))))))))..)))....)))))))..)))..)............. ( -36.10) >DroSec_CAF1 76858 120 + 1 ACUGCUGACGGCCAGUAAGCCCAACAGGCUCAUUCCAAUCACCUGGGCUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUA (((((....(((......)))(((((((...........((((((.....)))))).(((..((((((((.....))))))))..)))....)))))))............))))).... ( -40.10) >DroSim_CAF1 78446 120 + 1 ACUGCUGACGGCCAGUAAGCCCAACAGGCUCAUUCCAAUCACCUGGGAUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUA (((((....(((......)))(((((((...........((((((.....)))))).(((..((((((((.....))))))))..)))....)))))))............))))).... ( -40.10) >consensus ACUGCUGACGGCCAGUAAGCCCAACAGGCUCAUUCCAAUCACCUGGGAUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUA ..(((((.....)))))(((.(((((((...........((((((.....)))))).(((..((((((((.....))))))))..)))....)))))))..)))................ (-35.74 = -35.63 + -0.11)

| Location | 19,809,985 – 19,810,105 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -37.91 |
| Energy contribution | -38.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19809985 120 - 22407834 UAUAACUAUAUCUACGAACUGCAACAGGUAUUCGAGGGACUCAACAGCGAGCUGAGUCAAUCGGCACCUGAAUCCCAGGUGAUUGGAAUGAGCCUGCUGGGUUUACUGGGCGUUAGCACU ...................((((((((((..((((..((((((.(.....).))))))..))))((((((.....))))))..........))))(((.((....)).)))))).))).. ( -36.40) >DroSec_CAF1 76858 120 - 1 UAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAGCCCAGGUGAUUGGAAUGAGCCUGUUGGGCUUACUGGCCGUCAGCAGU .................((((((((((((((((((..((((((.(.....).))))))..))..((((((.....))))))....))))..))))))).((((....))))....))))) ( -43.80) >DroSim_CAF1 78446 120 - 1 UAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAUCCCAGGUGAUUGGAAUGAGCCUGUUGGGCUUACUGGCCGUCAGCAGU .................((((((((((((((((((..((((((.(.....).))))))..))..((((((.....))))))....))))..))))))).((((....))))....))))) ( -43.80) >consensus UAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAUCCCAGGUGAUUGGAAUGAGCCUGUUGGGCUUACUGGCCGUCAGCAGU .................((((((((((((((((((..((((((.(.....).))))))..))..((((((.....))))))....))))..))))))).((((....))))....))))) (-37.91 = -38.47 + 0.56)

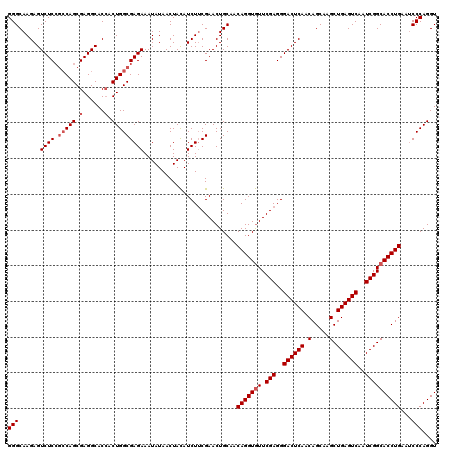
| Location | 19,810,025 – 19,810,145 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -39.27 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
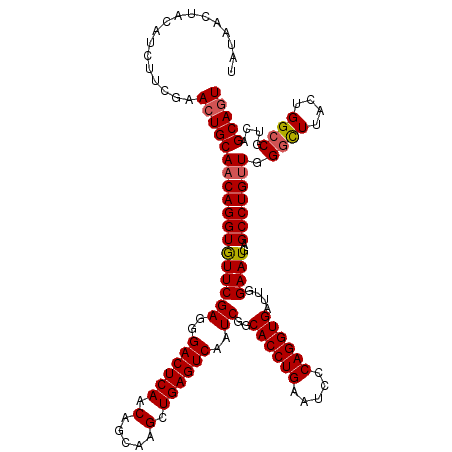
>2L_DroMel_CAF1 19810025 120 + 22407834 ACCUGGGAUUCAGGUGCCGAUUGACUCAGCUCGCUGUUGAGUCCCUCGAAUACCUGUUGCAGUUCGUAGAUAUAGUUAUAUUUCUCGCCAGUUGUGCCUCGCUGUUUGAGACUCUUGCCC ..(((((...((((((.(((..((((((((.....))))))))..)))..))))))...)....((.(((.(((....))).)))))))))......((((.....)))).......... ( -32.40) >DroSec_CAF1 76898 120 + 1 ACCUGGGCUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUAUUUCUCGCCAGUGGUGCCUCGCUGGCGGAGACUCUUGCCC ....((((..((((((.(((..((((((((.....))))))))..)))..))))))(((((.........))))).....((((.(((((((((....))))))))))))).....)))) ( -48.80) >DroSim_CAF1 78486 120 + 1 ACCUGGGAUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUAUUUCUCGCCAGUGGUGCCUCGCUGGCGGAGACUCUUGCCC ....(((...((((((.(((..((((((((.....))))))))..)))..))))))(((((.........))))).....((((.(((((((((....)))))))))))))......))) ( -45.50) >consensus ACCUGGGAUUCAGGUGCCGAUUGACUCAGCUUGCUGUUGAGUCCCUCGAACACCUGUUGCAGUUCGAAGAUGUAGUUAUAUUUCUCGCCAGUGGUGCCUCGCUGGCGGAGACUCUUGCCC ....(((...((((((.(((..((((((((.....))))))))..)))..))))))..........................((((((((((((....)))))))).))))......))) (-39.27 = -39.50 + 0.23)


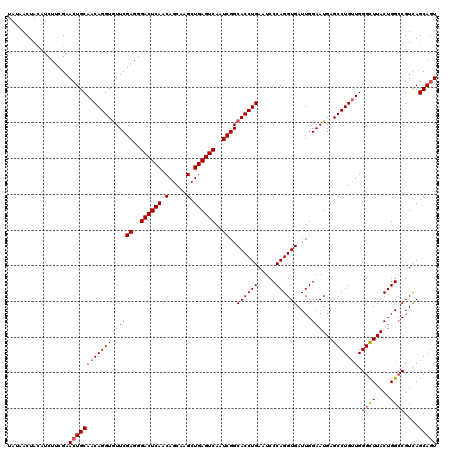
| Location | 19,810,025 – 19,810,145 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -35.33 |
| Energy contribution | -36.67 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19810025 120 - 22407834 GGGCAAGAGUCUCAAACAGCGAGGCACAACUGGCGAGAAAUAUAACUAUAUCUACGAACUGCAACAGGUAUUCGAGGGACUCAACAGCGAGCUGAGUCAAUCGGCACCUGAAUCCCAGGU (((...(((((((.....((.((......)).)).(((.(((....))).))).((((.(((.....))))))).)))))))..(((.(.(((((.....))))).))))...))).... ( -35.20) >DroSec_CAF1 76898 120 - 1 GGGCAAGAGUCUCCGCCAGCGAGGCACCACUGGCGAGAAAUAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAGCCCAGGU ((((..(((((((((((((.(......).)))))).)).............((((((((.((.....)))))))))))))))..(((...(((((.....)))))..)))..)))).... ( -47.30) >DroSim_CAF1 78486 120 - 1 GGGCAAGAGUCUCCGCCAGCGAGGCACCACUGGCGAGAAAUAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAUCCCAGGU (((...(((((((((((((.(......).)))))).)).............((((((((.((.....)))))))))))))))..(((...(((((.....)))))..)))...))).... ( -42.50) >consensus GGGCAAGAGUCUCCGCCAGCGAGGCACCACUGGCGAGAAAUAUAACUACAUCUUCGAACUGCAACAGGUGUUCGAGGGACUCAACAGCAAGCUGAGUCAAUCGGCACCUGAAUCCCAGGU (((......((((.(((((.(......).)))))))))..........................(((((((.(((..((((((.(.....).))))))..))))))))))...))).... (-35.33 = -36.67 + 1.33)

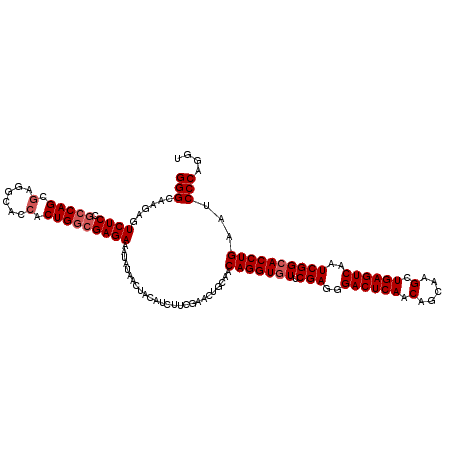
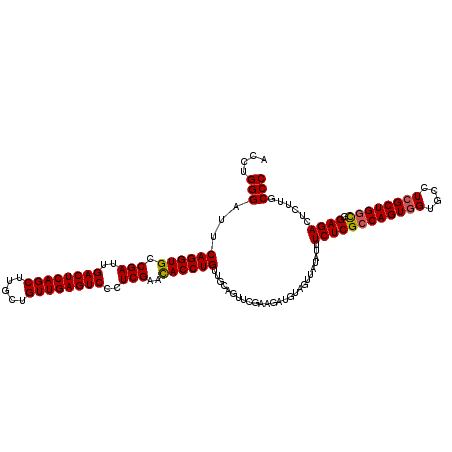
| Location | 19,810,145 – 19,810,265 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -32.31 |
| Energy contribution | -31.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19810145 120 - 22407834 UGGCAUUCAAAUCCAUAAUAUGAUGUCACUCACCAUCAAAUUCGUGCAACAAGUUGUGGACACAGUUCCGCUUGAACAACGUGGACCGGGAACGGCUGCCCUCCAGGCCUAUGCCAACAA ((((((.....((((((((.((.((.(((..............))))).)).)))))))).........(((((......(..(.(((....))))..)....)))))..)))))).... ( -33.94) >DroSec_CAF1 77018 120 - 1 UGGCAUUCAAAUCCAUAAUAUGAUGUCACUCACCAUCAAAUUCGUGCAGCAAGUUGUGGAUAGCGUUCCGGUUGAACAACGUGGACCGGGAACGGCUACCCUCCAGGCCUAUGCCGACAA .(((((....(((((((((.((.((.(((..............))))).)).)))))))))...(((((((((.(......).)))).)))))((((........)))).)))))..... ( -37.04) >DroSim_CAF1 78606 120 - 1 UGGCAUUCAAAUCCAUAAUAUGAUGUCACUCACCCUCAAAUUCGUGCAGCAAGUUGUGGACAGCGUUCCGAUUGAACAACGUGGACCGGGAACGGCUACCCUACAGGCCUAUGCCAACAA ((((((.....((((((((.((.((.(((..............))))).)).))))))))..((((((.....))))...((((.(((....))))))).......))..)))))).... ( -34.34) >consensus UGGCAUUCAAAUCCAUAAUAUGAUGUCACUCACCAUCAAAUUCGUGCAGCAAGUUGUGGACAGCGUUCCGAUUGAACAACGUGGACCGGGAACGGCUACCCUCCAGGCCUAUGCCAACAA ((((((.....((((((((.((.((.(((..............))))).)).))))))))....((((.....))))...((((.(((....)))))))...........)))))).... (-32.31 = -31.87 + -0.44)

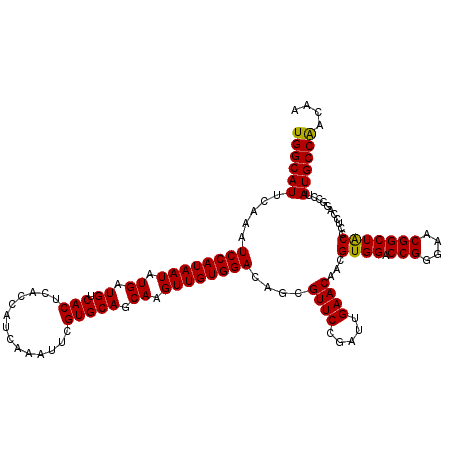
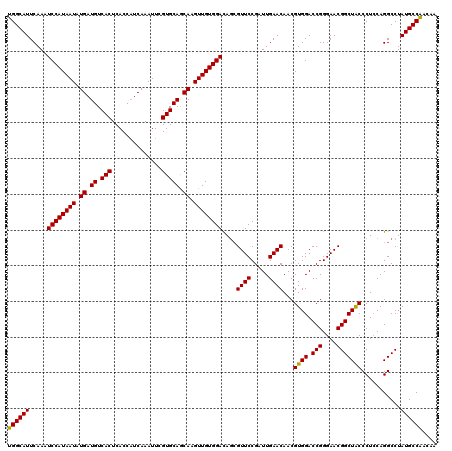
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:03 2006