
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,807,187 – 19,807,464 |
| Length | 277 |
| Max. P | 0.999993 |

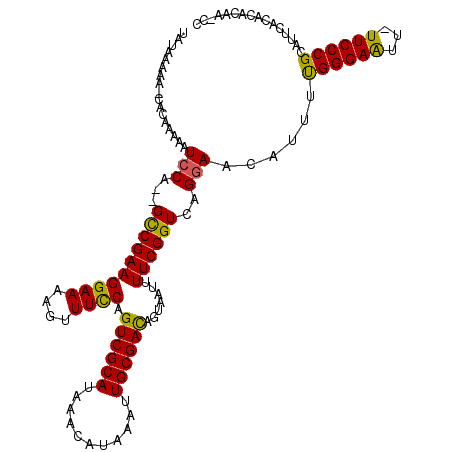
| Location | 19,807,187 – 19,807,304 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -26.52 |
| Energy contribution | -25.84 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.75 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19807187 117 - 22407834 UAUAAAAA-CACAAAAAUCCAGAGCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCAGGAACAUUUUGGGAAUU-UUCCCGCAUUCACACACAA-CC ........-........(((.((.(((((((((....)))).((((((...........)))))).......))))))).))).......((((...-.))))..............-.. ( -28.40) >DroSec_CAF1 73790 116 - 1 UAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCAGGAACAUUUCGGGAAUU-UUCCCGCAUUCACACACAUCCC ........-........(((.--((((((((((....)))).((((((...........)))))).......))))))..)))......(((((...-.)))))................ ( -29.50) >DroSim_CAF1 75573 116 - 1 UAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCAGGAACAUUUCGGGAAUU-UUCCCGCAUUCACACACAUCCC ........-........(((.--((((((((((....)))).((((((...........)))))).......))))))..)))......(((((...-.)))))................ ( -29.50) >DroEre_CAF1 75403 115 - 1 UAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCAGAAGUAUUUUGGGAAUU-UUCCCGCAUUCACACACAA-CC ........-........((..--((((((((((....)))).((((((...........)))))).......))))))..))........((((...-.))))..............-.. ( -26.40) >DroYak_CAF1 76920 117 - 1 UAUAAAAACCACAAAAAUCCA--GUCGAAGGAAAAGUUUUCAGUCGCAUAAACAUAAAUUGCGAUAGUAAUUUUCGGUCAGGAACAUUUUGGGAGUUUUUCCCGCAUUUACACACGA-CC .....................--((((..((((((..(..((((((((...........))))))......((((.....)))).....))..)..)))))).(......)...)))-). ( -20.90) >consensus UAUAAAAA_CACAAAAAUCCA__GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCAGGAACAUUUUGGGAAUU_UUCCCGCAUUCACACACAA_CC .................(((...((((((((((....)))).((((((...........)))))).......))))))..)))......((((((...))))))................ (-26.52 = -25.84 + -0.68)

| Location | 19,807,225 – 19,807,344 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -19.38 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19807225 119 - 22407834 UUUCCGUUUAUCCGAUUCUCUCCUCCUGUCGAACUUGAACUAUAAAAA-CACAAAAAUCCAGAGCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCA .....(((((..((((...........))))....)))))........-............((.(((((((((....)))).((((((...........)))))).......))))))). ( -24.00) >DroSec_CAF1 73829 117 - 1 CUUCCGUUUAUCUGAUUCUCUCCUCCUGUCGAACUUGAACUAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCA .....(((((..((((...........))))....)))))........-............--((((((((((....)))).((((((...........)))))).......)))))).. ( -22.00) >DroSim_CAF1 75612 117 - 1 UUUCCGUUUAUCUGAUUCUCUCCUCCUGUCGAACUUGAACUAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCA .....(((((..((((...........))))....)))))........-............--((((((((((....)))).((((((...........)))))).......)))))).. ( -22.00) >DroEre_CAF1 75441 117 - 1 UUUCCGUUUAUCUGGUUCUCUACUCGUGUCGAACUUGAACUAUAAAAA-CACAAAAAUCCA--GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCA .....((((.(.((((((.....(((...)))....)))))).).)))-)...........--((((((((((....)))).((((((...........)))))).......)))))).. ( -26.80) >DroYak_CAF1 76959 118 - 1 UUUCAGUUUAUCUGGUUUGCUCAUCCUGUCGAACUUGAACUAUAAAAACCACAAAAAUCCA--GUCGAAGGAAAAGUUUUCAGUCGCAUAAACAUAAAUUGCGAUAGUAAUUUUCGGUCA .....((((((((((...(((..((((.((((.((.((...................)).)--)))))))))..)))..))))....))))))..(((((((....)))))))....... ( -20.61) >consensus UUUCCGUUUAUCUGAUUCUCUCCUCCUGUCGAACUUGAACUAUAAAAA_CACAAAAAUCCA__GCCGAAGGAAAAGUUUCCAGUCGCAUAAACAUAAAUUGCGACAGUAAUUUUCGGUCA .....(((((..((((...........))))....))))).......................((((((((((....)))).((((((...........)))))).......)))))).. (-19.38 = -18.50 + -0.88)


| Location | 19,807,344 – 19,807,464 |
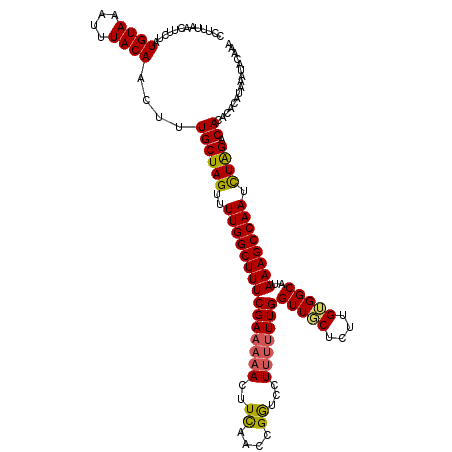
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -19.10 |
| Energy contribution | -18.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19807344 120 + 22407834 CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUCAACCGGUCCUUUUUUGGUUGCUCCUGUGGCAUAAAGCCAAUCUAGACACACACAUAAAUACCAA .............((((....))))....((((((..(((((((((((((((..((....))...)))))))((..(....)..))..)))))))).)))).))................ ( -21.70) >DroSec_CAF1 73946 120 + 1 CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUCAACCGGUCCUUUUUUGGUUGCUCUUGUGGCAUAAAGCCAAUCUAGACACAUACAUAAAUACAAA ...........((((((......((....))((((..(((((((((((((((..((....))...)))))))((..(....)..))..)))))))).)))).....))))))........ ( -22.30) >DroSim_CAF1 75729 120 + 1 CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUCAACCGGUCCUUUUUUGGUUGCUCCUGUGGCAUAAAGCCAAUCUAGACACACACAUAAGUACAAA ......((((...((((....))))....((((((..(((((((((((((((..((....))...)))))))((..(....)..))..)))))))).)))).)).......))))..... ( -22.60) >DroEre_CAF1 75558 119 + 1 CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUCACCCGAGC-UUUUUUGGUUCCUUUUGUGGCAUAAAGCCAAUUUGGACACAGACAUAAAUACAAA .............((((..((((...((.((((((..(((((((((((((((.(((....))).-)))))))((..(....)..))..)))))))).)))).)).))...)))))))).. ( -21.10) >DroYak_CAF1 77077 116 + 1 CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUUAACUGAGC-U---UUGGUUCCUCUUGGGGCAUAAAGCCAAUUUAGACACACACAUAAAUACAAA .............((((..((((......((((((..((((((((((((...(((.....))).-)---)))(((((....)))))..)))))))).)))).))......)))))))).. ( -19.30) >consensus CCUUUAACUUCUAUGUAAAUUUACAACUUUGCUAGUUUUGGCUUUCGAAAAACUUCAACCGGUCCUUUUUUGGUUGCUCUUGUGGCAUAAAGCCAAUCUAGACACACACAUAAAUACAAA .............((((....))))....((((((..(((((((((((((((..((....))...)))))))(((((....)))))..)))))))).)))).))................ (-19.10 = -18.86 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:48 2006