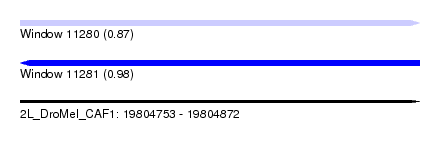
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,804,753 – 19,804,872 |
| Length | 119 |
| Max. P | 0.981529 |

| Location | 19,804,753 – 19,804,872 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -27.02 |
| Energy contribution | -26.42 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
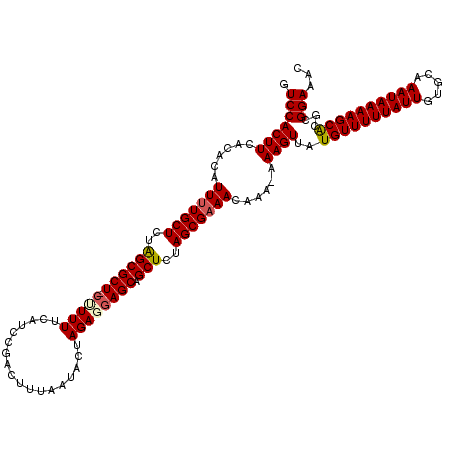
>2L_DroMel_CAF1 19804753 119 + 22407834 GUUUCCGCGUGCUUUUAUUUGCACAAUAAAAACAUAACUUU-UUUGUUUCGCUAGAGCUGCUCGUCUAGUAUUAAAGUCGGAUGAAAAAGAAGCGCCAGAGCAAAAUGUGUGAAGUGGAC ...(((((((((........))))........((((....(-(((((((((((....((..((((((..(......)..))))))...)).))))...))))))))..))))..))))). ( -31.20) >DroSec_CAF1 71397 118 + 1 GUUUCCGCGUGCUUUUAUUUGCACAAUAAAAACAUAACUUU-UUUGUUUCGCUGGAGCUGCUCCUCUAGUAUUAAAGUCGGAUGAAAAAG-AGCGCUAGAGCAAAAUGUGUGAAGUGGAC ...(((((((((........)))).......(((((.((((-((..(((.(((((((......))))))).........)))..))))))-.((......))....)))))...))))). ( -30.70) >DroSim_CAF1 72628 118 + 1 GUUUCCGCGUGCUUUUAUUUGCACAAUAAAAACAUAACUUU-UUUGUUUCGCUAGAGCUGCUCCUCUAGUAUUAAAGUCGGAUGAAAAAG-AGCGCUAGAGCAAAAUGUGUGAAGUGGAC ...(((((((((........)))).......(((((.((((-((..(((.(((((((......))))))).........)))..))))))-.((......))....)))))...))))). ( -31.00) >DroEre_CAF1 72816 119 + 1 GUUUCCGCGUGCUUUUAUUUGCACAAUAAAAACAUAACUUUUUUUGUUGCGCUGGAGCUGCUUCUCUAGUAUUAAAGUCGGAUGAAAAGG-AGCGCUAGAGCAAAAUGUGUGAAGUGGAC ...(((((((((........)))).......(((((..........((((.((((.(((.(((.((.................)).))).-))).)))).))))..)))))...))))). ( -31.33) >DroYak_CAF1 74363 119 + 1 GUUUCCGCGCGCUUUUAUUUGCACAAUAAAAACAUAACUUUUUUUGUUUCGCUAGAGCUGCUCUUCUAGCAUUAAAGUUAGAUGAAAAGG-AGCGCUAGAGCAAAAUGUGUGAAGUGGAC ...(((((.........((..((((....(((((..........))))).(((..(((.(((((((((((......))))))......))-))))))..)))....))))..))))))). ( -32.80) >consensus GUUUCCGCGUGCUUUUAUUUGCACAAUAAAAACAUAACUUU_UUUGUUUCGCUAGAGCUGCUCCUCUAGUAUUAAAGUCGGAUGAAAAAG_AGCGCUAGAGCAAAAUGUGUGAAGUGGAC ...((((((((((((((((((.((.....(((((..........))))).(((((((......)))))))......)))))))))).....)))))....(((.....)))...))))). (-27.02 = -26.42 + -0.60)

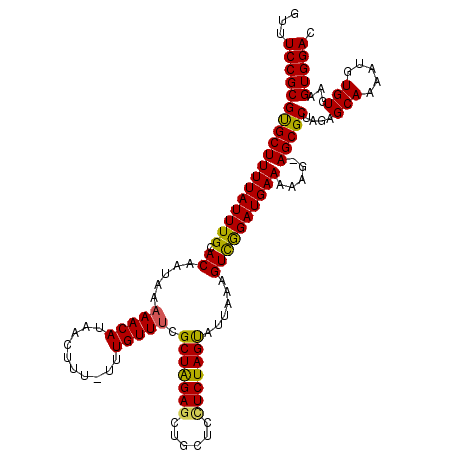
| Location | 19,804,753 – 19,804,872 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.79 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19804753 119 - 22407834 GUCCACUUCACACAUUUUGCUCUGGCGCUUCUUUUUCAUCCGACUUUAAUACUAGACGAGCAGCUCUAGCGAAACAAA-AAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCACGCGGAAAC .(((((((......(((((((..(((((((.(((...................))).)))).)))..)))))))....-.))))..((((((((((.....))))))))))...)))... ( -24.01) >DroSec_CAF1 71397 118 - 1 GUCCACUUCACACAUUUUGCUCUAGCGCU-CUUUUUCAUCCGACUUUAAUACUAGAGGAGCAGCUCCAGCGAAACAAA-AAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCACGCGGAAAC .(((((((......(((((((..((((((-(((((..................)))))))).)))..)))))))....-.))))..((((((((((.....))))))))))...)))... ( -29.27) >DroSim_CAF1 72628 118 - 1 GUCCACUUCACACAUUUUGCUCUAGCGCU-CUUUUUCAUCCGACUUUAAUACUAGAGGAGCAGCUCUAGCGAAACAAA-AAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCACGCGGAAAC .(((((((......(((((((..((((((-(((((..................)))))))).)))..)))))))....-.))))..((((((((((.....))))))))))...)))... ( -29.57) >DroEre_CAF1 72816 119 - 1 GUCCACUUCACACAUUUUGCUCUAGCGCU-CCUUUUCAUCCGACUUUAAUACUAGAGAAGCAGCUCCAGCGCAACAAAAAAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCACGCGGAAAC .(((............((((.((.(.(((-.((((((.................)))))).))).).)).))))........((..((((((((((.....)))))))))).)))))... ( -24.73) >DroYak_CAF1 74363 119 - 1 GUCCACUUCACACAUUUUGCUCUAGCGCU-CCUUUUCAUCUAACUUUAAUGCUAGAAGAGCAGCUCUAGCGAAACAAAAAAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCGCGCGGAAAC .(((((((......(((((((..((((((-(.(((.(((.........)))..))).)))).)))..)))))))......))))...(((((((((.....)))))))))....)))... ( -26.70) >consensus GUCCACUUCACACAUUUUGCUCUAGCGCU_CUUUUUCAUCCGACUUUAAUACUAGAGGAGCAGCUCUAGCGAAACAAA_AAAGUUAUGUUUUUAUUGUGCAAAUAAAAGCACGCGGAAAC .(((((((......(((((((..((((((.(((((..................)))))))).)))..)))))))......))))..((((((((((.....))))))))))...)))... (-23.39 = -23.79 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:40 2006