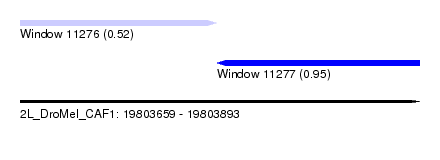
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,803,659 – 19,803,893 |
| Length | 234 |
| Max. P | 0.952048 |

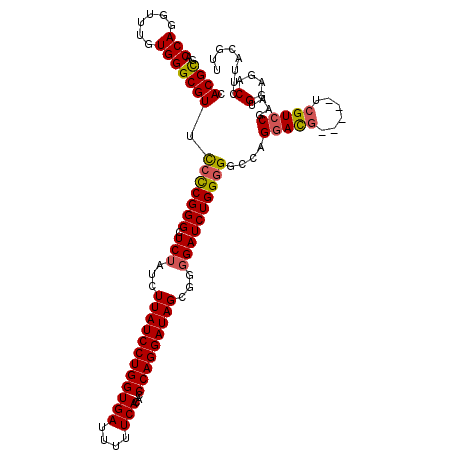
| Location | 19,803,659 – 19,803,774 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.34 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19803659 115 + 22407834 CACGCGACCAGGUUUGUGGGCGUUUCCCGGGCUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGGGGAUCUGGGGGACAGGACG-----UCGUCCAAAUGGAGACCUUACGUU ...(((...((((((.(((((((..((((((..((...((((((((((((....)))...)))))))))..))..))))))..))..(...-----.)))))).....))))))..))). ( -44.20) >DroSec_CAF1 70275 115 + 1 CACGCGACCAGGUUUGUGGGCGUUCCCCGGGCUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGGGGAUCUGGGGGCCAGGACG-----UCGUCCGAAUGGAGACCUUACGUU ...(((...((((((.((..((..(((((((..((...((((((((((((....)))...)))))))))..))..)))))))..).((((.-----..)))))..)).))))))..))). ( -44.70) >DroSim_CAF1 71506 115 + 1 CACGCGACCAGGUUUGUGGGCGUUCCCCGGGCUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGGGGAUCUGGGGGCCAGGACG-----UCGUCCGAAUGGAGACCUUACGUU ...(((...((((((.((..((..(((((((..((...((((((((((((....)))...)))))))))..))..)))))))..).((((.-----..)))))..)).))))))..))). ( -44.70) >DroEre_CAF1 71722 120 + 1 CACGCGACCAAAUUUGUGGGCGUUCCUCGGGCUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGGGGAUCUGGGGGCCAGAAUGCAAUGGAGUCCGAAUGGAGACCUUACGUU ...............(((((..(((((((((((((((..(((((((((((....)))...))))))))(((....(((((...))))).))).)))))))))))..))))..)))))... ( -44.70) >DroYak_CAF1 73327 114 + 1 CACGUGACCAAAUUUGUGGGCGUACACCGGGUUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGG-GAUCUGGGGGCCAGGACG-----UCGUCCAAAUGGAGACCUUGCGUU .(((..(......((.(((.(.....(((((((((...((((((((((((....)))...))))))))).))-))))))).).))).)).(-----((.(((....)))))).)..))). ( -39.30) >consensus CACGCGACCAGGUUUGUGGGCGUUCCCCGGGCUCUAUCUUAUCCUGGUGAUUUUUCACACCCAGGAUAGCGGGGAUCUGGGGGCCAGGACG_____UCGUCCGAAUGGAGACCUUACGUU .((((..(((......))))))).(((((((.(((...((((((((((((....)))...)))))))))...))))))))))....(((((......)))))....(....)........ (-36.18 = -36.34 + 0.16)


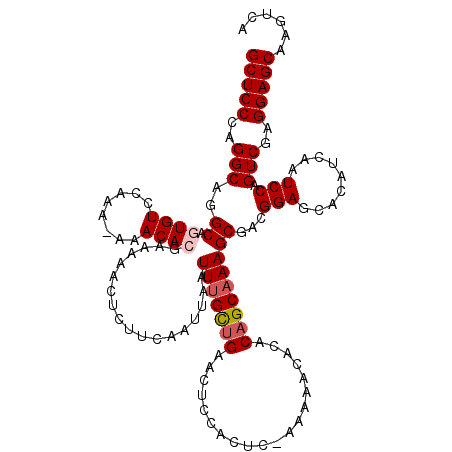
| Location | 19,803,774 – 19,803,893 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.79 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19803774 119 - 22407834 GCUCCCAGGCAGGCAGUGUCCAAA-AAACACGAAGAACUCUUCAAUUAAUUCGCUGAACUCCACUCUAAAAACACACAGCAAAGCGACGGAGCACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..(((.((..((((((...-......((((....)))).......(((((...........................))))).))).)))......)).)))..)))))...... ( -28.63) >DroSec_CAF1 70390 119 - 1 GCUCCCAGGCAGGCAGUGUCCAAA-AAACACGAAAAACUCUUCAAUUAAUUUGCUGAACUCCACUCGAAAAACACACAGCAAAGCGACGGAGCACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..(((..((.((((.....-..))))..................(((((((....................)))))))))...(((.........))).)))..)))))...... ( -27.45) >DroSim_CAF1 71621 119 - 1 GCUCCCAGGCAGGCAGUGUCCAAA-AAACACGGAAAACUCUUCAAUUAAUUUGCCGAACUCCACUCGAAAAACACACAGCAAAGCGACGGAGCACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..(((.((((((((.....-..)))).(((.....)))........))))...((((..(((.....(.....).....))).))))............)))..)))))...... ( -28.20) >DroEre_CAF1 71842 109 - 1 GCUCCCAGGCUGGCAUUGUCCAAA-AAACACGAAA-------CAAUUAAUUUGCUGAGCUCUGCUC---AGGCAAACAGCAAAGCGACGGAGCACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..((((((.((((......-..........-------.......((((((((((...))))---).))))).......((......))....))))))))))..)))))...... ( -35.30) >DroYak_CAF1 73441 116 - 1 GCUCCCAGGCUGGCAUUGUCCAAAAAAACACGAAA-ACUCUUCAAUUAAUUUGUUGAGCUCUGCUC---AAACAAACAGCAAAGCGACGGAGAACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..((((((.(((((((..............-.....(((((......)))))(((.((((.---........)))).)))...)))......))))))))))..)))))...... ( -34.80) >consensus GCUCCCAGGCAGGCAGUGUCCAAA_AAACACGAAAAACUCUUCAAUUAAUUUGCUGAACUCCACUC_AAAAACACACAGCAAAGCGACGGAGCACAUCAAUCCAGUCGAGGAGCAAGUCA (((((..(((..((.((((........))))..................(((((((....................)))))))))...(((.........))).)))..)))))...... (-23.15 = -23.79 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:36 2006