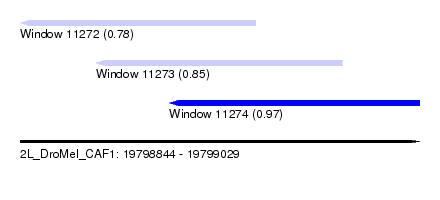
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,798,844 – 19,799,029 |
| Length | 185 |
| Max. P | 0.969754 |

| Location | 19,798,844 – 19,798,953 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.18 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.63 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
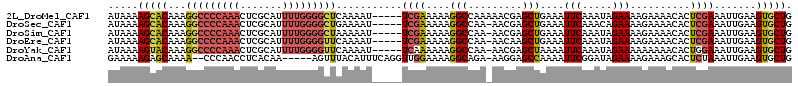
>2L_DroMel_CAF1 19798844 109 - 22407834 AUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCAAAAU-----UCGAAAAAGGCCAAAAACGAGCUGAAAUUCAAAUAGAAAAGAAAACACUCGAAAUUGAAGUGCUG .....(((((....((((((((.......))))))))((((..(-----((((....((((......).)))....(((.....)))..........))))).)))).))))). ( -27.50) >DroSec_CAF1 65409 108 - 1 AUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUGAAAAU-----UCGAAAAAGGCCAA-AACGAGCUGAAAUUCAAACAGAAAAGAAAACACUCGAAAUUGAAGUGCUG .....(((((...(((((((((.......))))))))).....(-----((((....(((...-.....)))....(((.....)))..........)))))......))))). ( -27.40) >DroSim_CAF1 66652 108 - 1 AUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUAAAAAU-----UCGAAAAAGGCCAA-AACGAGCUGAAAUUCAAAUAGAAAAGAAAACACUCGAAAUUGAAGUGCUG .....(((((...(((((((((.......))))))))).....(-----((((....(((...-.....)))....(((.....)))..........)))))......))))). ( -27.40) >DroEre_CAF1 66807 108 - 1 AUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGUUCAAAAU-----UCGAAAAAGGCCAA-AACAAGCUGAAAUUCAAAUAGAAAAGAAAACACUCGAAAUUGAAGUGCUG .....(((((....((((((((.......))))))))((((..(-----((((....(((...-.....)))....(((.....)))..........))))).)))).))))). ( -25.30) >DroYak_CAF1 67775 108 - 1 AUAAAAGUACAAAGGCCCCAAACUCGCAUUUUGGGGUUCAAAAU-----UCAAAAAAGGCCAA-AACGAGCUAAAAUUCAAAUAGAAAAAAAAACACUGGAAAUUGAAGUGCUG .....(((((....((((((((.......))))))))((((..(-----(((.....(((...-.....)))....(((.....)))..........))))..)))).))))). ( -21.20) >DroAna_CAF1 64586 106 - 1 GAAAAAGAGCAAAA--CCCAACCUCACAA-----AGUUUACAUUUCAGGUUGGAAAAGGCAGA-AAGGAGCCAAAAUUCGGAUAGAAAAGAAAGCACUCUAAAUUGAAGUGCUG (((...........--.(((((((.....-----............)))))))....(((...-.....)))....))).............(((((((......).)))))). ( -21.43) >consensus AUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCAAAAU_____UCGAAAAAGGCCAA_AACGAGCUGAAAUUCAAAUAGAAAAGAAAACACUCGAAAUUGAAGUGCUG .....(((((...(((((((((.......)))))))))...........((((....(((.........)))....(((.....)))..........)))).......))))). (-17.63 = -18.63 + 1.00)
| Location | 19,798,879 – 19,798,993 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -13.49 |
| Energy contribution | -15.10 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19798879 114 - 22407834 CAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCAAAAU-----UCGAAAAAGGCCAAAAACGAGCUGAAAUUC ..(((((.......)))))........(((..((............)).....(((((((((.......)))))))))......-----....................)))....... ( -23.50) >DroSec_CAF1 65444 111 - 1 CAGGUGUACA--UAGCACCAAUAAUAGGGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUGAAAAU-----UCGAAAAAGGCCAA-AACGAGCUGAAAUUC ..(((((...--..)))))........(((..((............)).....(((((((((.......)))))))))......-----.........)))..-............... ( -26.60) >DroSim_CAF1 66687 113 - 1 CAGGUGUACAAGUAGCACCAAUAACAAGGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUAAAAAU-----UCGAAAAAGGCCAA-AACGAGCUGAAAUUC ..(((((.......)))))........(((..((............)).....(((((((((.......)))))))))......-----.........)))..-............... ( -26.50) >DroEre_CAF1 66842 113 - 1 CAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGUUCAAAAU-----UCGAAAAAGGCCAA-AACAAGCUGAAAUUC ..(((((.......)))))........(((..((............)).....((((......(((.((((((.....))))))-----.)))....))))..-.....)))....... ( -22.90) >DroYak_CAF1 67810 113 - 1 CAGGUGUACAAAUAGCGCCAAUAAUAAAGCAAGCAUAACCAUAAAAGUACAAAGGCCCCAAACUCGCAUUUUGGGGUUCAAAAU-----UCAAAAAAGGCCAA-AACGAGCUAAAAUUC ..(((((.......)))))............(((...................(((((((((.......)))))))))......-----........(.....-..)..)))....... ( -18.90) >DroAna_CAF1 64621 99 - 1 CAGAUGUUUAAA------------GAAAACAAACAAAAUUGAAAAAGAGCAAAA--CCCAACCUCACAA-----AGUUUACAUUUCAGGUUGGAAAAGGCAGA-AAGGAGCCAAAAUUC ....(((((...------------..))))).......................--.(((((((.....-----............)))))))....(((...-.....)))....... ( -14.43) >consensus CAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCAAAAU_____UCGAAAAAGGCCAA_AACGAGCUGAAAUUC ..(((((.......)))))..................................(((((((((.......)))))))))...................(((.........)))....... (-13.49 = -15.10 + 1.61)
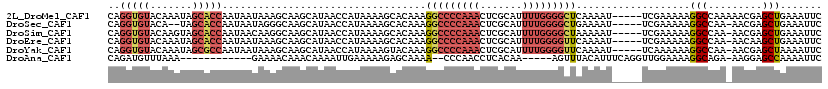
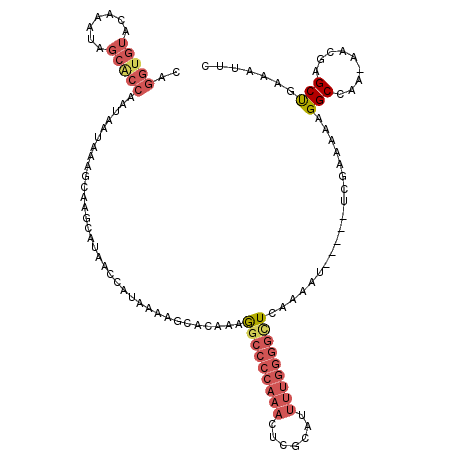
| Location | 19,798,913 – 19,799,029 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -12.23 |
| Energy contribution | -13.98 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
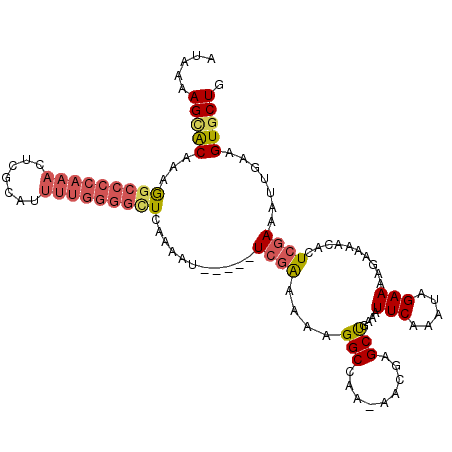
>2L_DroMel_CAF1 19798913 116 - 22407834 AGCUAAAUG-ACCUAAAAACUGGAAAGG---GUAUCGAAACAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCA .(((.....-.((........))....(---((.........(((((.......))))).........(....)...))).....))).....(((((((((.......))))))))).. ( -26.50) >DroSec_CAF1 65477 114 - 1 AGCUAAAGG-UCCUAAAAAUUCGAAAGG---GUAUUGAAACAGGUGUACA--UAGCACCAAUAAUAGGGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUGA .(((....(-(((((...((((....))---)).........(((((...--..))))).....)))))).)))...................(((((((((.......))))))))).. ( -35.70) >DroSim_CAF1 66720 116 - 1 AGCUAGAGG-ACCUGAAAAUUCGAAAGG---GUAUUGAAACAGGUGUACAAGUAGCACCAAUAACAAGGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUAA .(((....(-.(((....((((....))---)).........(((((.......))))).......)))).)))...................(((((((((.......))))))))).. ( -29.60) >DroEre_CAF1 66875 115 - 1 AACUAAAUG-ACC-AGAGUGUCCAGAGA---GUAUCGAAACAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGUUCA .........-...-...((((...((..---...))......(((((.......)))))...........................))))...(((((((((.......))))))))).. ( -21.40) >DroYak_CAF1 67843 116 - 1 AACUAAAUGGACC-AGAGUGUCCAGAGA---GUAUCGAAACAGGUGUACAAAUAGCGCCAAUAAUAAAGCAAGCAUAACCAUAAAAGUACAAAGGCCCCAAACUCGCAUUUUGGGGUUCA .(((...(((((.-.....)))))...)---)).........(((((.......)))))..................................(((((((((.......))))))))).. ( -26.20) >DroAna_CAF1 64659 100 - 1 AGUUUUAAG-GAAGAAAACCUUCAAAGGACAAUAUCAUACCAGAUGUUUAAA------------GAAAACAAACAAAAUUGAAAAAGAGCAAAA--CCCAACCUCACAA-----AGUUUA .(((((...-((((.....))))..)))))((((((......))))))....------------......((((....((....))(((.....--......)))....-----.)))). ( -9.80) >consensus AGCUAAAUG_ACCUAAAAACUCCAAAGG___GUAUCGAAACAGGUGUACAAAUAGCACCAAUAAUAAAGCAAGCAUAACCAUAAAAGCACAAAGGCCCCAAACUCGCAUUUUGGGGCUCA ..........................................(((((.......)))))..................................(((((((((.......))))))))).. (-12.23 = -13.98 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:34 2006