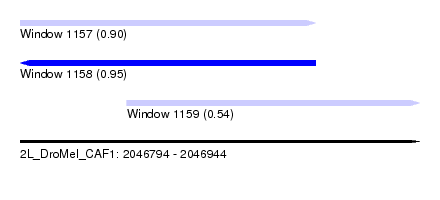
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,046,794 – 2,046,944 |
| Length | 150 |
| Max. P | 0.950687 |

| Location | 2,046,794 – 2,046,905 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -17.05 |
| Energy contribution | -16.08 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
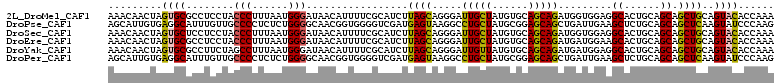
>2L_DroMel_CAF1 2046794 111 + 22407834 AAACAACUAGUGCGCCUCCUACCCUUUAAUGGGAUAACAUUUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGGUGGAGGCACUGCAGCAGCUGCAGUACACCAAA .........(((.((((((..(((......)))....(((((.(((((..((((((...)))))).)))).).)))))..))))))(((((((...))))))).))).... ( -40.10) >DroPse_CAF1 82895 111 + 1 AGCAUUGUGAGGCAUUUGUUGCCCCUCUCUGGGGCAACGGUGGGGUCGAUGAGUAAGGCCUGCUAUGCGGAGCAGCUGAUUGAAGCUCUGCAGCAGCUCAAGUAUCCCAAG .((........))..((((((((((.....))))))))))(((((((..((((......(((((..(((((((..(.....)..)))))))))))))))).).)))))).. ( -46.50) >DroSec_CAF1 64254 111 + 1 AAACAACUAGUGCUCCUCCUACCCUUUAAUGGGAUAACAUUUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGGUGGAGGCACUGCAGCAGCUGCAGUACACCAAA .........(((..(((((..(((......)))....(((((.(((((..((((((...)))))).)))).).)))))..))))).(((((((...))))))).))).... ( -35.20) >DroEre_CAF1 63144 111 + 1 AAACAACUAGUGCGCCUCCUACCCUUUAAUGGGAUAACAUUUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGAUGGAAGCACUGCAGCAGCUGCAGUACACCAAA .........(((.((.(((..(((......)))....(((((.(((((..((((((...)))))).)))).).)))))..))).))(((((((...))))))).))).... ( -33.40) >DroYak_CAF1 66465 111 + 1 AAACAACUAGUGCGCCUUCUAGCCUUUAAUGGGAUAACAUUUUCGCAUCUUAGCAGGGAUUGUUAUGUGCAGCAGAUGAUGGAGGCACUGCAGCAGCUGCAGUACACCAAA .........(((.((((((...(((.....)))....(((((.(((((..((((((...)))))).)))).).)))))..))))))(((((((...))))))).))).... ( -31.70) >DroPer_CAF1 95237 111 + 1 AGCAUUGUGAGGCAUUUGUUGCCCCUCUCUGGGGCAACGGUGGGGUCGAUGAGUAAGGCCUGCUAUGCGGAGCAGCUGAUUGAAGCUCUGCAGCAGCUCAAGUAUCCCAAG .((........))..((((((((((.....))))))))))(((((((..((((......(((((..(((((((..(.....)..)))))))))))))))).).)))))).. ( -46.50) >consensus AAACAACUAGUGCGCCUCCUACCCUUUAAUGGGAUAACAUUUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGAUGGAAGCACUGCAGCAGCUGCAGUACACCAAA .........((((........(((......))).................((((.....(((((......))))).........((......)).))))..))))...... (-17.05 = -16.08 + -0.97)
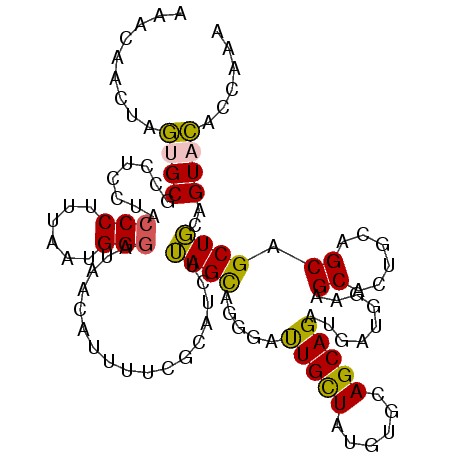
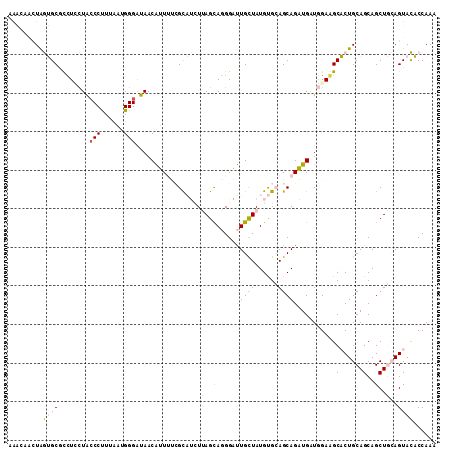
| Location | 2,046,794 – 2,046,905 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.63 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2046794 111 - 22407834 UUUGGUGUACUGCAGCUGCUGCAGUGCCUCCACCAUCUGCUGCACAUAGCAAUCCCUGCUAAGAUGCGAAAAUGUUAUCCCAUUAAAGGGUAGGAGGCGCACUAGUUGUUU .((((((((((((((...)))))))(((((((((......(((((.(((((.....))))).).))))..((((......))))....))).)))))))))))))...... ( -41.10) >DroPse_CAF1 82895 111 - 1 CUUGGGAUACUUGAGCUGCUGCAGAGCUUCAAUCAGCUGCUCCGCAUAGCAGGCCUUACUCAUCGACCCCACCGUUGCCCCAGAGAGGGGCAACAAAUGCCUCACAAUGCU ..((((.....(((((((((((.((((..(.....)..)))).))..)))))......)))).....))))..((((((((.....))))))))................. ( -37.90) >DroSec_CAF1 64254 111 - 1 UUUGGUGUACUGCAGCUGCUGCAGUGCCUCCACCAUCUGCUGCACAUAGCAAUCCCUGCUAAGAUGCGAAAAUGUUAUCCCAUUAAAGGGUAGGAGGAGCACUAGUUGUUU .((((((((((((((...))))))).((((((((......(((((.(((((.....))))).).))))..((((......))))....))).))))).)))))))...... ( -37.30) >DroEre_CAF1 63144 111 - 1 UUUGGUGUACUGCAGCUGCUGCAGUGCUUCCAUCAUCUGCUGCACAUAGCAAUCCCUGCUAAGAUGCGAAAAUGUUAUCCCAUUAAAGGGUAGGAGGCGCACUAGUUGUUU .((((((((((((((...)))))))((((((.........(((((.(((((.....))))).).))))..........(((......)))..)))))))))))))...... ( -38.00) >DroYak_CAF1 66465 111 - 1 UUUGGUGUACUGCAGCUGCUGCAGUGCCUCCAUCAUCUGCUGCACAUAACAAUCCCUGCUAAGAUGCGAAAAUGUUAUCCCAUUAAAGGCUAGAAGGCGCACUAGUUGUUU ..(((.(((((((((...)))))))))..))).((.(((.(((..((((((.....(((......)))....))))))..........(((....)))))).))).))... ( -29.10) >DroPer_CAF1 95237 111 - 1 CUUGGGAUACUUGAGCUGCUGCAGAGCUUCAAUCAGCUGCUCCGCAUAGCAGGCCUUACUCAUCGACCCCACCGUUGCCCCAGAGAGGGGCAACAAAUGCCUCACAAUGCU ..((((.....(((((((((((.((((..(.....)..)))).))..)))))......)))).....))))..((((((((.....))))))))................. ( -37.90) >consensus UUUGGUGUACUGCAGCUGCUGCAGUGCCUCCAUCAUCUGCUGCACAUAGCAAUCCCUGCUAAGAUGCGAAAAUGUUAUCCCAUUAAAGGGUAGGAGGCGCACUAGUUGUUU ..((((((.((((((...)))))).((((........(((((....))))).......................((((((.......)))))).))))))))))....... (-18.82 = -18.63 + -0.19)

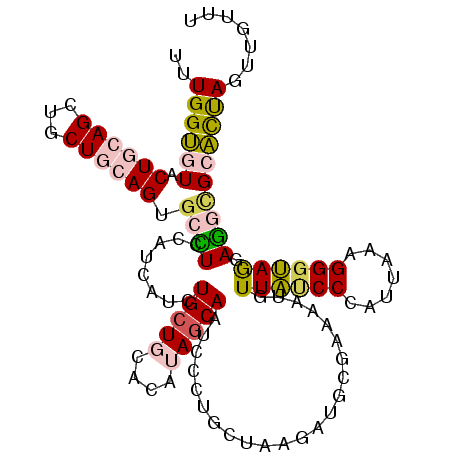
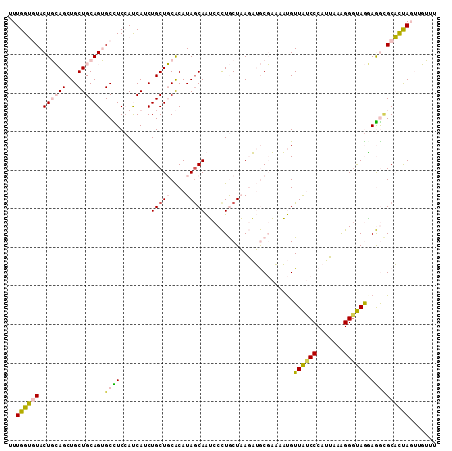
| Location | 2,046,834 – 2,046,944 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -20.81 |
| Energy contribution | -19.32 |
| Covariance contribution | -1.49 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2046834 110 + 22407834 UUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGGUGGAGGCACUGCAGCAGCUGCAGUACACCAAAAAGGUGAACAAAAGCUGGCUUCAAACGCCGAACAGCACA- ....((((..((((((...)))))).)))).((...(((((((((((((((((...))))))).((((.....))))...........)))))...)))))....))...- ( -36.20) >DroPse_CAF1 82935 110 + 1 UGGGGUCGAUGAGUAAGGCCUGCUAUGCGGAGCAGCUGAUUGAAGCUCUGCAGCAGCUCAAGUAUCCCAAGAAGGUGAACAGGACCUGGCUGUUGAUGCCGGG-AACACAG .((.((((((.(((.(((((((((.((((((((..(.....)..))))))))..)))....((.((((.....)).))))))).))).))))))))).))(..-..).... ( -40.40) >DroSim_CAF1 62687 110 + 1 UUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGGUGGAGGCACUGCAGCAGCUGCAGUACACCAAAAAGGUGAACAAAAGCUGGCUUCAAACGCCGAACAGCACA- ....((((..((((((...)))))).)))).((...(((((((((((((((((...))))))).((((.....))))...........)))))...)))))....))...- ( -36.20) >DroEre_CAF1 63184 110 + 1 UUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGAUGGAAGCACUGCAGCAGCUGCAGUACACCAAAAAGGUGAACAAAAGUUGGCUUCAAACGCCGAACAGUGCA- ((((((((((..(((.(........).)))...))))).)))))((((((..((....)).((.((((.....)))).)).....(((((.......))))).)))))).- ( -33.10) >DroYak_CAF1 66505 110 + 1 UUUCGCAUCUUAGCAGGGAUUGUUAUGUGCAGCAGAUGAUGGAGGCACUGCAGCAGCUGCAGUACACCAAAAAGGUGAACAAAAGCUGGCUUCAAACGCCGAACAGCACA- ....((((..((((((...)))))).)))).((..........((((((((((...))))))).((((.....)))).......)))(((.......))).....))...- ( -30.70) >DroPer_CAF1 95277 110 + 1 UGGGGUCGAUGAGUAAGGCCUGCUAUGCGGAGCAGCUGAUUGAAGCUCUGCAGCAGCUCAAGUAUCCCAAGAAGGUGAACAGGACCUGGCUGUUGAUGCCGGG-AACACAG .((.((((((.(((.(((((((((.((((((((..(.....)..))))))))..)))....((.((((.....)).))))))).))).))))))))).))(..-..).... ( -40.40) >consensus UUUCGCAUCUUAGCAGGGAUUGCUAUGUGCAGCAGAUGAUGGAAGCACUGCAGCAGCUGCAGUACACCAAAAAGGUGAACAAAAGCUGGCUUCAAACGCCGAACAGCACA_ ....((....((((.....(((((......))))).........((......)).))))..((.((((.....)))).))......((((.......))))....)).... (-20.81 = -19.32 + -1.49)
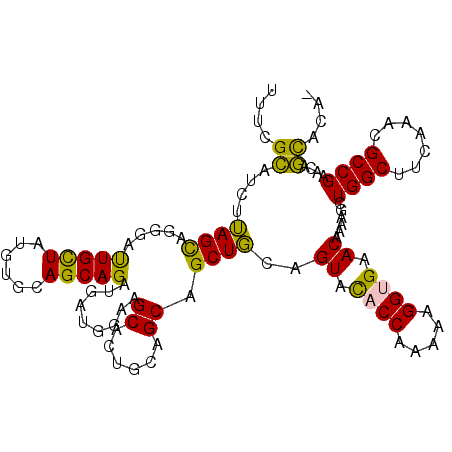
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:28 2006