| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,797,150 – 19,797,265 |
| Length | 115 |
| Max. P | 0.546408 |

| Location | 19,797,150 – 19,797,265 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19797150 115 + 22407834 ACCCCUCC---GCUCGCAGCUUCCUUCUCUCGACUGAGCAAGAGUUUGGCUACAUUUGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU ..((((..---....((((((.((..(((......))).((((((.((((.......)))).)))))).))(((..(.(((((.....))))).)..)))))))))....)))).... ( -34.60) >DroSec_CAF1 63686 104 + 1 ACCCCUCC---GC-----------UUCUCUCCGCUGAGCAAGAGUUUGGCUACUUUUGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU ..(((((.---((-----------........)).))((((((((......)))))))).......(((((((.((..(((((.....)))))((....)))).)))))))))).... ( -31.50) >DroSim_CAF1 64932 104 + 1 ACCCCUCC---GC-----------UUCUCUCCGCUGAGCAAGAGUUUGGCUACUUUUGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU ..(((((.---((-----------........)).))((((((((......)))))))).......(((((((.((..(((((.....)))))((....)))).)))))))))).... ( -31.50) >DroEre_CAF1 65022 115 + 1 ACCCCUCU---GCUCGCAGCUUCCUUCGCUCGGCUUAGCAAAAGUUUGGCUACAUUUGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU ..((((.(---(...((((((.((...(((......)))((((((.((((.......)))).)))))).))(((..(.(((((.....))))).)..))))))))).)).)))).... ( -36.90) >DroYak_CAF1 66019 115 + 1 ACCCCUCU---GCUCGUAGCUUCCUUCUCUCGGCUUAGCAAGAGUUUGGCUACAUUCGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU .(((((((---(((...((((..........)))).))).))))..((((.......)))).....(((((((.((..(((((.....)))))((....)))).)))))))))).... ( -34.40) >DroAna_CAF1 63018 118 + 1 UCAUCUCAACGGCUGGGGGACCUUUCCACACGUUUUCGAAUGAGUUUUGUUACAUUUGGUGUAUUUUUUUGGUUGCCUAGGAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU .....(((((..(((((.((((.....((((.....((((((..........))))))))))........)))).)))))((((......)))))))))......(((...))).... ( -26.32) >consensus ACCCCUCC___GCUCG_AGCUUCCUUCUCUCGGCUGAGCAAGAGUUUGGCUACAUUUGCCAUAUUUUUUGGGUUGCCUAGCAGUUUAUCUGCUUGUUGACAGCUGCCCAAAGGGAAUU ..((((..................................(((((.((((.......)))).)))))((((((.((..(((((.....)))))((....)))).)))))))))).... (-22.79 = -23.23 + 0.45)

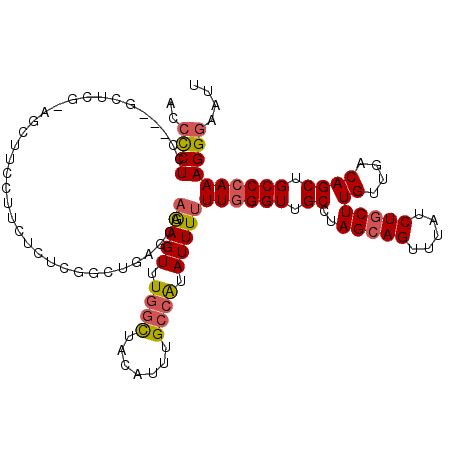
| Location | 19,797,150 – 19,797,265 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -21.68 |
| Energy contribution | -22.93 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19797150 115 - 22407834 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCAAAUGUAGCCAAACUCUUGCUCAGUCGAGAGAAGGAAGCUGCGAGC---GGAGGGGU ...((((((((.(((((((((....(((((.....))))).)))...((........((((.......))))..((((((......))))))..)).))))))...)---))))))). ( -39.90) >DroSec_CAF1 63686 104 - 1 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCAAAAGUAGCCAAACUCUUGCUCAGCGGAGAGAA-----------GC---GGAGGGGU ...(((((((((((.(((((((...(((((.....))))).(....)..........))))...))).)))...(((((((...)).)))))..-----------.)---))))))). ( -31.70) >DroSim_CAF1 64932 104 - 1 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCAAAAGUAGCCAAACUCUUGCUCAGCGGAGAGAA-----------GC---GGAGGGGU ...(((((((((((.(((((((...(((((.....))))).(....)..........))))...))).)))...(((((((...)).)))))..-----------.)---))))))). ( -31.70) >DroEre_CAF1 65022 115 - 1 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCAAAUGUAGCCAAACUUUUGCUAAGCCGAGCGAAGGAAGCUGCGAGC---AGAGGGGU ...((((((((.(((((((((....(((((.....))))).))).............((((.......))))..((((((((......)))))))).))))))...)---))))))). ( -40.90) >DroYak_CAF1 66019 115 - 1 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCGAAUGUAGCCAAACUCUUGCUAAGCCGAGAGAAGGAAGCUACGAGC---AGAGGGGU ...(((((((((((((.((((........))))..))))).(((..((.........((((.......))))..((((((......))))))..))..))).....)---))))))). ( -36.40) >DroAna_CAF1 63018 118 - 1 AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUCCUAGGCAACCAAAAAAAUACACCAAAUGUAACAAAACUCAUUCGAAAACGUGUGGAAAGGUCCCCCAGCCGUUGAGAUGA ......(((..(((.(((.......))).............((..(((.......((((.....)))).......((((.((....)).))))...)))..))..)))...))).... ( -18.00) >consensus AAUUCCCUUUGGGCAGCUGUCAACAAGCAGAUAAACUGCUAGGCAACCCAAAAAAUAUGGCAAAUGUAGCCAAACUCUUGCUAAGCCGAGAGAAGGAAGCU_CGAGC___GGAGGGGU ...((((((((((..(((.......(((((.....))))).)))..)))).......((((.......))))..(((((........)))))...................)))))). (-21.68 = -22.93 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:30 2006