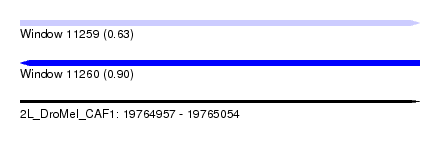
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,764,957 – 19,765,054 |
| Length | 97 |
| Max. P | 0.903273 |

| Location | 19,764,957 – 19,765,054 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -19.44 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19764957 97 + 22407834 --------ACCGCGCGCGCGCUUAUGUUGUUUGCUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUUGCUCUAAACGAGCUGC---------------C --------...(((....))).......((..(((((((.......(.(((((...(((((......)))))..))))))(((.....)))...))))))).))---------------. ( -29.70) >DroVir_CAF1 33703 105 + 1 CCGUUGCCGCUGCCGUCGCGCUUAUGUUGUUUACUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUCGCUCUAAGCGAUGCUC---------------C ..((((..((.(((((((((..((.......))..)).))))))).))(((((...(((((......)))))..)))))))))..(((((.....)))))....---------------. ( -36.70) >DroGri_CAF1 31720 97 + 1 --------GCUGCCGUCGCGCGUAGGUUGUUUACUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUCGCUCUAAGCGAUGUCC---------------C --------((.(((((((((.((((.....)))).)).))))))).))(((((...(((((......)))))..)))))......(((((.....)))))....---------------. ( -36.70) >DroWil_CAF1 41090 100 + 1 --------GCGA----CAAUUUGAUGUUGUUUACUCGUUGACGGUCGCGUUGAGUUUGUGGCAAUCACCGCACAUCAACCAGCAAAUCGCUCUAAACGAGCAGA-GC-------GCUUCC --------((((----(...(..(((.........)))..)..)))))(((((...(((((......)))))..))))).(((...((((((.....)))).))-..-------)))... ( -30.30) >DroMoj_CAF1 35241 94 + 1 -----------GCCGUCGCGCUUAUGUUGUUUGCUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUCGCUAUAAGCGAUGUCC---------------C -----------...(.(((((((((((.(((((((((....))...(.(((((...(((((......)))))..))))))))))))).)).))))))).)).).---------------. ( -31.80) >DroAna_CAF1 30344 108 + 1 --------GCUGC----GCGCUUAUGUUGUUUGCUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUUGCUCCAAACGAGCAGCAGCACCAGCAGCAUCC --------(((((----.......(((((((.(((((((...((..(.(((((...(((((......)))))..))))))(((.....))))).))))))))))))))...))))).... ( -42.80) >consensus ________GCUGCCGUCGCGCUUAUGUUGUUUACUCGUUGACGGUCGCGUUGAGUUUGCGGCAAUCACCGCACAUCAACCAGCAAAUCGCUCUAAACGAGCUGC_______________C ..........................(((((((...((.....((.(.(((((...(((((......)))))..)))))).)).....))..)))))))..................... (-19.44 = -18.67 + -0.78)

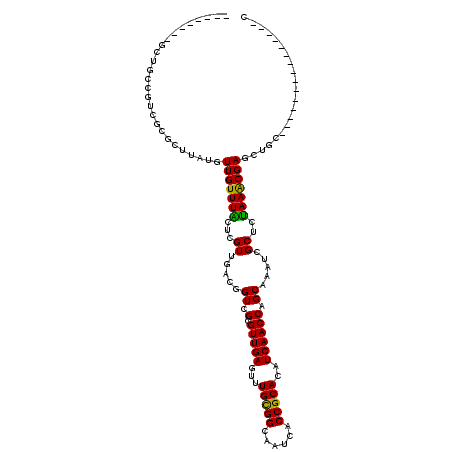
| Location | 19,764,957 – 19,765,054 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19764957 97 - 22407834 G---------------GCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCGCGCGCGGU-------- .---------------((.((.((((((......((((((.(((((((((((((....))))))...((.....)).))))))).)))))).....)))))).))..))...-------- ( -37.00) >DroVir_CAF1 33703 105 - 1 G---------------GAGCAUCGCUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGUAAACAACAUAAGCGCGACGGCAGCGGCAACGG .---------------.((((((((.....))))..)))).(((((..((((((....))))))...)))))((..(((((..((..((.......))..)).)))))..))(....).. ( -37.90) >DroGri_CAF1 31720 97 - 1 G---------------GGACAUCGCUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGUAAACAACCUACGCGCGACGGCAGC-------- (---------------(.(.(((((.....))))).).)).(((((..((((((....))))))...)))))((..(((((..((.(((.......))).)).)))))..))-------- ( -35.20) >DroWil_CAF1 41090 100 - 1 GGAAGC-------GC-UCUGCUCGUUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCACAAACUCAACGCGACCGUCAACGAGUAAACAACAUCAAAUUG----UCGC-------- .((..(-------((-((((......))))))).((((((.(((((((((((.(((((......)).))).))))..))))))).)))))).....))......----....-------- ( -30.20) >DroMoj_CAF1 35241 94 - 1 G---------------GGACAUCGCUUAUAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCGACGGC----------- .---------------......(((((((...(.((((((.(((((((((((((....))))))...((.....)).))))))).)))))))...))))))).......----------- ( -32.20) >DroAna_CAF1 30344 108 - 1 GGAUGCUGCUGGUGCUGCUGCUCGUUUGGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGC----GCAGC-------- ....(((((..(((((..(((((.....))))).((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......)))))----)))))-------- ( -45.00) >consensus G_______________GCACAUCGCUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCGACGGCAGC________ ......................((((((......((((((.(((((((((((((....))))))...((.....)).))))))).)))))).....)))))).................. (-26.56 = -26.62 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:21 2006