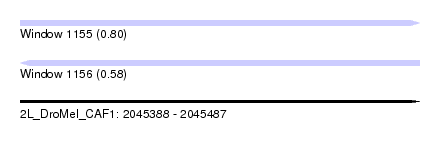
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,045,388 – 2,045,487 |
| Length | 99 |
| Max. P | 0.795113 |

| Location | 2,045,388 – 2,045,487 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -17.80 |
| Energy contribution | -19.12 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
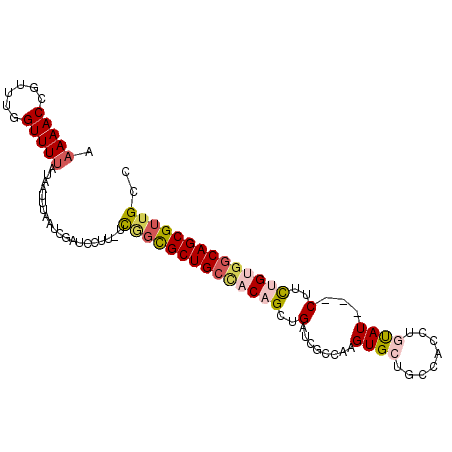
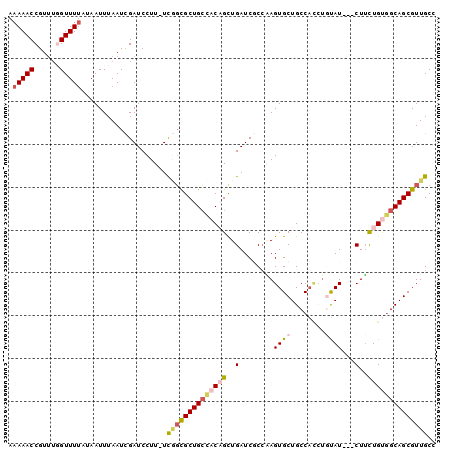
>2L_DroMel_CAF1 2045388 99 + 22407834 AAAAACCGUUUGGUUUUAUAAUUUAAUCGAUCCUU-UCCCUGCUGCCACAGCUGAUCGCCGAGUGCUGCCACUUGUAUCCUCUUCUGUGGCAGCGUUGCC .((((((....))))))..................-.....((((((((((..((....((((((....)))))).....))..))))))))))...... ( -31.40) >DroSec_CAF1 62845 96 + 1 AAAAACCGUAUGGUUUUAAAAUUUAAUUGAUCCUU-UCAGCGCUGACACAGCUGAUCUCAAAGUGCUGCCAUCUGUAU---CUUCUGUGGCAGCGUUGCC .((((((....))))))..................-.((((((((.(((((..(((..((..(((....))).)).))---)..))))).)))))))).. ( -26.00) >DroSim_CAF1 61241 96 + 1 AAAAACCGUUUGGUUUUAAAAUUUAAUUGAUCCUU-UCGGCGCUGCUACAGCUGAUCUCCAAGUGCUGCCAUCUGUAU---CUUCUGUGGCAGCGUUGCC .((((((....))))))..................-.((((((((((((((..(((...((..((....))..)).))---)..)))))))))))))).. ( -30.90) >DroEre_CAF1 61737 96 + 1 AAAAACCGUUGCGUUUUAUAAUUUAAUCGAUCCUU-UCGGCGCUGCCACUGUUGAUCGCCCAGUGCUGCCACUUCUAU---CUGCUGAUGCAGCGUUGCC ......(((((((((.............((.....-))((((..((.((((.........))))))))))........---.....)))))))))..... ( -20.60) >DroYak_CAF1 65047 92 + 1 AAAAACCGUUUCGUUUCAUAAUUUAAUCGAUCCUUUUUGGCGCUGCCACUGUUGAUCGCCAAGUGCUGCC-----CAU---CUUUUGCGGCAGCGUUGCC ...........................(((((..(..((((...))))..)..))))).(((.(((((((-----((.---....)).)))))))))).. ( -23.40) >consensus AAAAACCGUUUGGUUUUAUAAUUUAAUCGAUCCUU_UCGGCGCUGCCACAGCUGAUCGCCAAGUGCUGCCACCUGUAU___CUUCUGUGGCAGCGUUGCC .(((((......)))))....................((((((((((((((..(........((((........))))...)..)))))))))))))).. (-17.80 = -19.12 + 1.32)

| Location | 2,045,388 – 2,045,487 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -16.85 |
| Energy contribution | -18.05 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
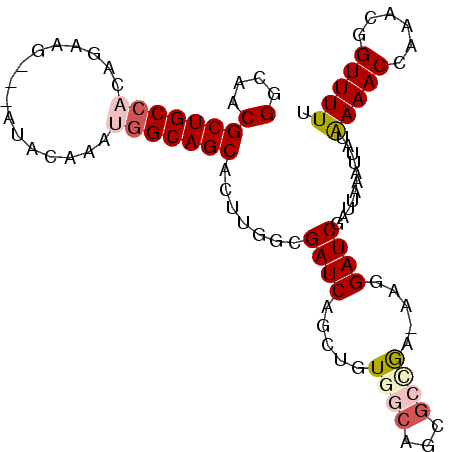
>2L_DroMel_CAF1 2045388 99 - 22407834 GGCAACGCUGCCACAGAAGAGGAUACAAGUGGCAGCACUCGGCGAUCAGCUGUGGCAGCAGGGA-AAGGAUCGAUUAAAUUAUAAAACCAAACGGUUUUU (....)((((((((((.....(((.(.((((....)))).)...)))..)))))))))).....-..................((((((....)))))). ( -31.60) >DroSec_CAF1 62845 96 - 1 GGCAACGCUGCCACAGAAG---AUACAGAUGGCAGCACUUUGAGAUCAGCUGUGUCAGCGCUGA-AAGGAUCAAUUAAAUUUUAAAACCAUACGGUUUUU (....)((((((((.....---.....).))))))).((((.((....((((...)))).)).)-)))...............((((((....)))))). ( -25.70) >DroSim_CAF1 61241 96 - 1 GGCAACGCUGCCACAGAAG---AUACAGAUGGCAGCACUUGGAGAUCAGCUGUAGCAGCGCCGA-AAGGAUCAAUUAAAUUUUAAAACCAAACGGUUUUU .....((((((.((((..(---((.(((.((....)).)))...)))..)))).))))))((..-..))..............((((((....)))))). ( -28.50) >DroEre_CAF1 61737 96 - 1 GGCAACGCUGCAUCAGCAG---AUAGAAGUGGCAGCACUGGGCGAUCAACAGUGGCAGCGCCGA-AAGGAUCGAUUAAAUUAUAAAACGCAACGGUUUUU .((..((((((....((..---.........))..(((((.........)))))))))))((..-..))...................)).......... ( -24.20) >DroYak_CAF1 65047 92 - 1 GGCAACGCUGCCGCAAAAG---AUG-----GGCAGCACUUGGCGAUCAACAGUGGCAGCGCCAAAAAGGAUCGAUUAAAUUAUGAAACGAAACGGUUUUU (....)((((((.((....---.))-----))))))..(((((((((.....((((...)))).....))))).)))).....(((((......))))). ( -26.80) >consensus GGCAACGCUGCCACAGAAG___AUACAAAUGGCAGCACUUGGCGAUCAGCUGUGGCAGCGCCGA_AAGGAUCGAUUAAAUUAUAAAACCAAACGGUUUUU (....)(((((((................))))))).......((((.....((((...)))).....))))...........(((((......))))). (-16.85 = -18.05 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:25 2006