| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,725,327 – 19,725,422 |
| Length | 95 |
| Max. P | 0.707403 |

| Location | 19,725,327 – 19,725,422 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -17.71 |
| Energy contribution | -17.21 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19725327 95 + 22407834 ACUUUUGGCGCAUGCUCUGGGAGCACAACUCCACCAUUGUGGUCAUGCUGACCAAGCUCAAGGAAAUGGGAAGGGUAAGC-----UC--------CCCCCUUGCA---CCA .....(((.(((......((((((...((((..(((((.(((((.....)))))..........)))))...))))..))-----))--------))....))).---))) ( -32.40) >DroVir_CAF1 16399 94 + 1 ACUUUUGGCGCAUGCUCUGGGAGCACAACUCCACCAUAGUUGUUAUGUUGACCAAACUCAAGGAAAUGGGCAGGGUAAGC-----UG--------A-CA--UAUUCU-CCU ....(..((...(((((((((((.....)))).((((..((.((..(((.....)))..)).)).)))).))))))).))-----..--------)-..--......-... ( -23.50) >DroGri_CAF1 15296 106 + 1 ACUUUUGGCGCAUGCUCUGGGAGCACAACUCCACCAUAGUUGUUAUGCUAACCAAACUCAAGGAAAUGGGCAGGGUAAGU-----UUCACAGCUCAACACUCGCUCCCUUU ......(((....)))..((((((((((((.......))))))...(((.(((...((((......))))...))).)))-----.................))))))... ( -25.30) >DroWil_CAF1 19588 97 + 1 ACUUUUGGCGCAUGCUCUGGGAACAUAAUUCCACCAUUGUAGUCAUGUUGACCAAACUAAAGGAAAUGGGCAGAGUAAGUGUAGU-----------CCACUUGUA---CAU .......((((.(((((((((((.....)))).(((((........(((.....))).......))))).))))))).))))...-----------.........---... ( -23.56) >DroMoj_CAF1 16406 96 + 1 ACUUCUGGCGCAUGCUCUGGGAGCACAACUCCACCAUAGUUGUUAUGUUAACCAAGCUCAAGGAAAUGGGCAGGGUAAGC-----UG--------AACA--UGCUGUAUAU .......(((((((.((..((((.....))))..............(((.(((..(((((......)))))..))).)))-----.)--------).))--))).)).... ( -25.10) >DroAna_CAF1 14185 93 + 1 ACUUUUGGCGAAUGCUCUGGGAGCAUAACUCCACCAUUGUGGUGAUGCUCACCAAGCUCAAGGAAAUGGGCAGGGUAUGU-----GC--------UGAUC--AAA---AGA .(((((((((.(((((((((((((((....((((....))))..)))))).))..(((((......)))))))))))).)-----))--------....)--)))---)). ( -36.60) >consensus ACUUUUGGCGCAUGCUCUGGGAGCACAACUCCACCAUAGUUGUUAUGCUGACCAAACUCAAGGAAAUGGGCAGGGUAAGC_____UC________AACAC_UGCA___CAU .........((.(((((((((((.....)))).((((.........(((.....)))........)))).))))))).))............................... (-17.71 = -17.21 + -0.50)

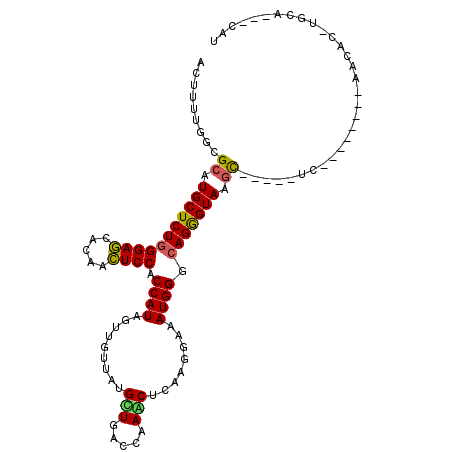

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:12 2006