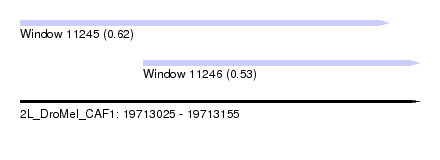
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,713,025 – 19,713,155 |
| Length | 130 |
| Max. P | 0.624137 |

| Location | 19,713,025 – 19,713,145 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.32 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
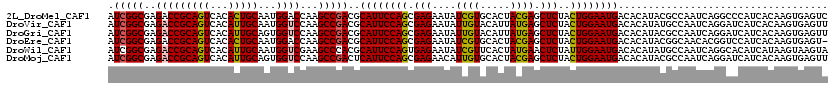
>2L_DroMel_CAF1 19713025 120 + 22407834 AUCGGCGAGACCGCAGUCACACUGCAAUGGACCAAGCCGACGCAUUCCAGCGAGAAUAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGCCCAUCACAAGUGAGUC .(((((....((((((.....))))...)).....))))).(((((((((.(((....(((((...))))).)))..)))))))).)......(((.....)))...(((....)))... ( -36.30) >DroVir_CAF1 2886 120 + 1 AUCGGCGAGACCGCAGUCACAUUGCAAUGGUCCAAGCCGACGCAUUCCAGCGAGAAUAUUGUACAUUAUGAGCUCUACUGGAAUGACACAUAUGCCAAUCAGGAUCAUCACAAGUGAGUU .(((((..(((((((((...)))))...))))...))))).(((((((((.(((.(((........)))...)))..)))))))).).......((.....)).((((.....))))... ( -32.50) >DroGri_CAF1 2409 120 + 1 AUCGGCGAGACCGCAGUCACAUUGCAGUGGUCCAAGCCGACGCAUUCCAGCGAGAAUAUUGUACAUUAUGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGAUCAUCACAAGUGAGUU .(((((..((((((.((......)).))))))...))))).(((((((((.(((.(((........)))...)))..)))))))).).......((.....)).((((.....))))... ( -34.30) >DroEre_CAF1 2329 119 + 1 AUCGGCGAGACCGCAGUCACACUGCAAUGGACCAAGCCGACGCAUUCCAGCGAGAAUAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGGCAACACGGUCCAUCACAAGUGAGU- ....(((....))).....((((...(((((((..((((...((((((((.(((....(((((...))))).)))..)))))))).......)))).....)))))))....))))...- ( -40.80) >DroWil_CAF1 2551 120 + 1 AUCGGCGAGACCGCAGUCACAUUGCAAUGGUCGAAGCCCACGCAUUCCAGUGAGAAUAUCGUUCACUAUGAACUCUAUUGGAAUGACACAUAUGCCAAUCAGGCACAUCAUAAGUAAGUA ...(((..(((((((((...)))))...))))...)))...(((((((((((((....((((.....)))).))).))))))))).).....((((.....))))............... ( -33.40) >DroMoj_CAF1 2952 120 + 1 AUCGGCGAGACCGCAGUCACAUUGCAGUGGUCCAAGCCGACUCAUUCCAGCGAGAACAUUGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGAUCAUCACAAGUGAGUU .(((((..((((((.((......)).))))))...))))).(((((((((.(((....(((((...))))).)))..)))))))))........((.....)).((((.....))))... ( -36.40) >consensus AUCGGCGAGACCGCAGUCACAUUGCAAUGGUCCAAGCCGACGCAUUCCAGCGAGAAUAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGAUCAUCACAAGUGAGUU .(((((..(((((((((...)))))...))))...)))))..((((((((.(((....((((.....)))).)))..))))))))................................... (-29.68 = -29.32 + -0.36)

| Location | 19,713,065 – 19,713,155 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19713065 90 + 22407834 CGCAUUCCAGCGAGAAUAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGCCCAUCACAAGUGAGUCCUU-AGUCUCG .(((((((((.(((....(((((...))))).)))..)))))))).)......(((.....)))...........(((.(...-.).))). ( -23.60) >DroPse_CAF1 2090 83 + 1 CGCAUUCCAGCGAGAACAUUGUUCACUACGAGCUCUACUGGAAUGACACAUAUGCCAAUCAGGCACAUCACAAGUGAGUAUUU-------- .(((((((((.(((....((((.....)))).)))..)))))))).).....((((.....))))..(((....)))......-------- ( -21.90) >DroEre_CAF1 2369 79 + 1 CGCAUUCCAGCGAGAAUAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGGCAACACGGUCCAUCACAAGUGAGU------------ ..((((((((.(((....(((((...))))).)))..)))))))).(((....(....)..(........)..)))...------------ ( -20.60) >DroMoj_CAF1 2992 81 + 1 CUCAUUCCAGCGAGAACAUUGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGAUCAUCACAAGUGAGUUA---------- .(((((((((.(((....(((((...))))).)))..)))))))))........((.....)).((((.....))))....---------- ( -18.00) >DroAna_CAF1 2061 91 + 1 CGCAUUCCAGCGAGAAUAUCGUACACUACGAGCUCUACUGGUAUGACACAUACGCCAAUCAGACCCAUCACAAGUGAGUUCUAGAGUCCCC .........((((.....))))...(((.((((((.(((.(((((...)))))........((....))...))))))))))))....... ( -16.60) >DroPer_CAF1 2015 83 + 1 CGCAUUCCAGCGAGAACAUUGUACACUACGAGCUCUACUGGAAUGACACAUAUGCCAAUCAGGCACAUCACAAGUGAGUAUUU-------- .(((((((((.(((....(((((...))))).)))..)))))))).).....((((.....))))..(((....)))......-------- ( -21.80) >consensus CGCAUUCCAGCGAGAACAUCGUGCACUACGAGCUCUACUGGAAUGACACAUACGCCAAUCAGGCCCAUCACAAGUGAGUACUU________ ..((((((((.(((....((((.....)))).)))..))))))))........(((.....)))...(((....))).............. (-17.01 = -17.32 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:09 2006