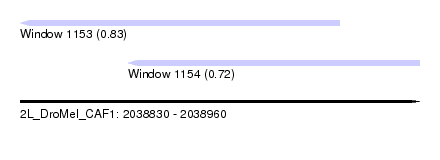
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,038,830 – 2,038,960 |
| Length | 130 |
| Max. P | 0.825206 |

| Location | 2,038,830 – 2,038,934 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.76 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.825206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
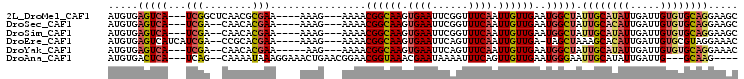
>2L_DroMel_CAF1 2038830 104 - 22407834 AAGAAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGCAACAUGGCCGAUAUGGCGAUACACAAGCCCACAAA----- .......((((((.((((......)))).))))))...(((((((((((((((.(..(((((.....)).)))..).)))))).))))).....))))......----- ( -28.00) >DroSec_CAF1 56248 104 - 1 AAGAAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCACAUUGAUUGUGUGCAGGAAGCAACAUGGCCGAUAUGGCGAUACACAAGCCAAGAAA----- .......((((((.((((......)))).))))))..(((((.((((((((.....))))))))........((.(((.....))).)).....))))).....----- ( -30.30) >DroSim_CAF1 54709 104 - 1 AAGAAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGCAACAUGGCCGAUAUGGCGAUACACAAGCCAAGAAA----- .......((((((.((((......)))).))))))..((((((((((((((((.(..(((((.....)).)))..).)))))).))))).....))))).....----- ( -29.90) >DroEre_CAF1 55036 98 - 1 AAGAAAACGGCAAGUGAAUUCAGUUUCAAUUGUUGA-UAGCUAAAGCACAUUGAUUGUGCGUAGGAAAC---------GAUAUAGCGAUACACAAACCAAGAAACACA- .......((((((.((((......)))).)))))).-..((((..(((((.....)))))...(....)---------....))))......................- ( -22.00) >DroYak_CAF1 58393 109 - 1 AAGAAAACGGCAAGUGAAUUCAGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAACAACAUGGUCGAUAUGGCGAUACACAAACCAAGAAAUACAA .......((((((.((((......)))).))))))..((..((((((((((((((..(((...(....).)))..)))))))).))))))..))............... ( -31.40) >consensus AAGAAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGCAACAUGGCCGAUAUGGCGAUACACAAGCCAAGAAA_____ .......((((((.((((......)))).))))))..((((((((((((((.....)))))))(....)...)))))))...((((.........)))).......... (-18.88 = -20.76 + 1.88)

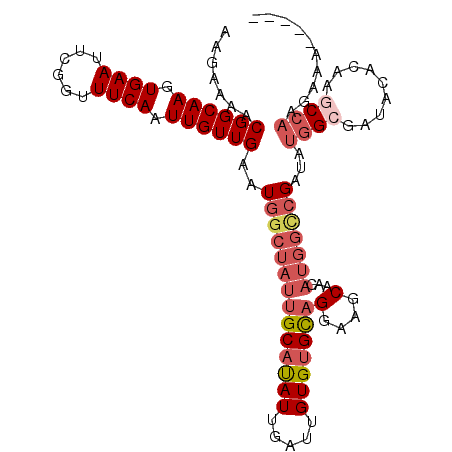
| Location | 2,038,865 – 2,038,960 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2038865 95 - 22407834 AUGUGAGUCA---UCGGCUCAACGCGAA----AAAG---AAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGC ...(((((..---...))))).......----....---....((((((.((((......)))).))))))....(((.((((((((.....))))))))..))) ( -24.30) >DroSec_CAF1 56283 93 - 1 AUGUGAGUCA---UCGA--CAACACGAA----AAAG---AAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCACAUUGAUUGUGUGCAGGAAGC .....(((((---((((--(((...(((----(..(---((.((.....))...)))..))))..)))))).)))))).((((((((.....))))))))..... ( -26.60) >DroSim_CAF1 54744 93 - 1 AUGUGAGUCA---UCGA--CAACACGAA----AAAG---AAAACGGCAAGUGAAUUCGGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGC .....(((((---((((--(((...(((----(..(---((.((.....))...)))..))))..)))))).)))))).((((((((.....))))))))..... ( -24.70) >DroEre_CAF1 55066 95 - 1 AUGUGAGUCAUCAUCGA--CCGCACGAA----AAAG---AAAACGGCAAGUGAAUUCAGUUUCAAUUGUUGA-UAGCUAAAGCACAUUGAUUGUGCGUAGGAAAC .((((.(((......))--)))))....----....---....((((((.((((......)))).)))))).-...(((..(((((.....))))).)))..... ( -22.60) >DroYak_CAF1 58433 92 - 1 AUGUGAGUCA---UCGA--CAACACGAA-----AAG---AAAACGGCAAGUGAAUUCAGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAAC .....(((((---((((--(((...(((-----(.(---((.((.....))...)))..))))..)))))).)))))).((((((((.....))))))))..... ( -22.10) >DroAna_CAF1 53315 93 - 1 AUGUGACUCA---UCAG--CAAAAUAAAGGAAACUGAACGGAACGGUAAACGAAUAAAAUUUCAGUUGUUGAAUGGGAAUUGCAUAUUGAUUG---GCAAG---- (((..(((((---((((--(((.....((....))................(((......)))..)))))).)))))..)..)))........---.....---- ( -16.90) >consensus AUGUGAGUCA___UCGA__CAACACGAA____AAAG___AAAACGGCAAGUGAAUUCAGUUUCAAUUGUUGAAUGGCUAUUGCAUAUUGAUUGUGUGCAGGAAGC .....(((((...(((........)))................((((((.((((......)))).))))))..))))).((((((((.....))))))))..... (-15.42 = -15.98 + 0.56)
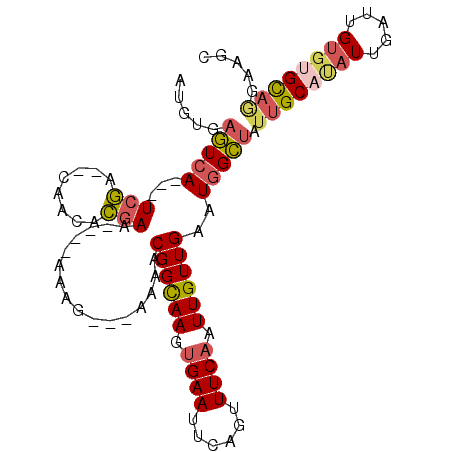
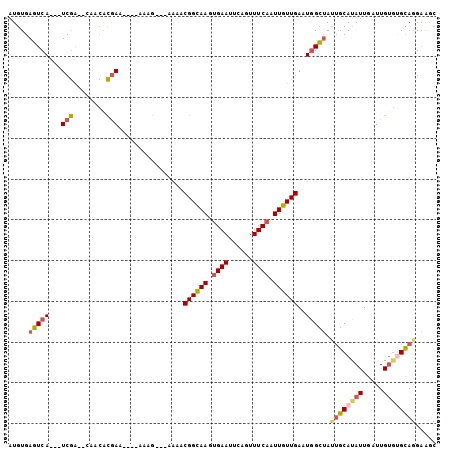
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:23 2006