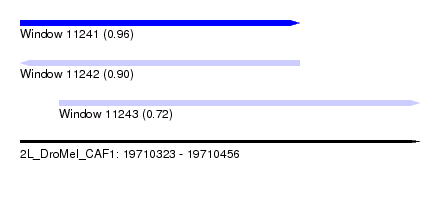
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,710,323 – 19,710,456 |
| Length | 133 |
| Max. P | 0.957813 |

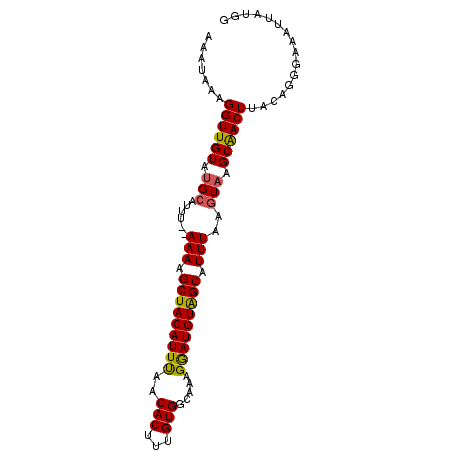
| Location | 19,710,323 – 19,710,416 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -16.77 |
| Energy contribution | -16.52 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19710323 93 + 22407834 AAAUAAAGGUUGUGUGCAUUUAUAAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUGUGG ......(((((((.(((.....((((.(((((((((..(((...))).....))))))))).)))).))).)))))))((((.....)))).. ( -19.90) >DroSim_CAF1 3682 91 + 1 AAAUAAAGGUUGUGUGCAUUU--AAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUGUGG ......(((((((....((((--(((.(((((((((..(((...))).....))))))))).)))))))..)))))))((((.....)))).. ( -22.40) >DroEre_CAF1 3634 86 + 1 AAAUAAAGGUUGUAAGCAUUU--AAAAGCUACAUUCAACACCACGUGGCAAAGGAUGUGGCAUUUAAAUAAGCGACU-----GCAAAUUAUGG .......((((((....((((--(((.(((((((((....((....))....))))))))).)))))))..))))))-----........... ( -22.40) >DroYak_CAF1 3705 91 + 1 AAAUAAUGGUUGUAUGCAUUU--AAAAGCUACAUUUAACACUUUGUGGCGAAGAAUGUAGCAUUUAGGUAAGCAACUUACAGGGAAAUUAUGG .......((((((.(((...(--(((.(((((((((..(.((....)).)..))))))))).)))).))).))))))................ ( -21.10) >consensus AAAUAAAGGUUGUAUGCAUUU__AAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUAUGG .......((((((.(((......(((.(((((((((..(((...))).....))))))))).)))..))).))))))................ (-16.77 = -16.52 + -0.25)

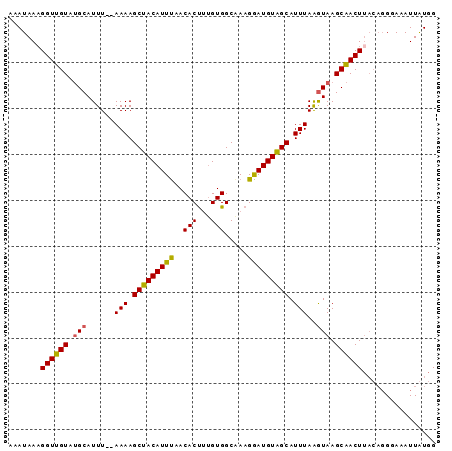
| Location | 19,710,323 – 19,710,416 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -15.18 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19710323 93 - 22407834 CCACAAUUUCCCUGUAAGUUGCUUACUUAAAUGCUACAUCCUUUGCCACAAAGUGUUAAAUGUAGCUUUUAUAAAUGCACACAACCUUUAUUU .................(((((.....((((.(((((((((((((...))))).)....))))))).)))).....))).))........... ( -14.80) >DroSim_CAF1 3682 91 - 1 CCACAAUUUCCCUGUAAGUUGCUUACUUAAAUGCUACAUCCUUUGCCACAAAGUGUUAAAUGUAGCUUUU--AAAUGCACACAACCUUUAUUU .................(((((....(((((.(((((((((((((...))))).)....))))))).)))--))..))).))........... ( -14.50) >DroEre_CAF1 3634 86 - 1 CCAUAAUUUGC-----AGUCGCUUAUUUAAAUGCCACAUCCUUUGCCACGUGGUGUUGAAUGUAGCUUUU--AAAUGCUUACAACCUUUAUUU .........((-----....)).((((((((.((.((((.(..((((....))))..).)))).)).)))--)))))................ ( -14.80) >DroYak_CAF1 3705 91 - 1 CCAUAAUUUCCCUGUAAGUUGCUUACCUAAAUGCUACAUUCUUCGCCACAAAGUGUUAAAUGUAGCUUUU--AAAUGCAUACAACCAUUAUUU ............((((...(((.....((((.((((((((...(((......)))...)))))))).)))--)...))))))).......... ( -16.60) >consensus CCACAAUUUCCCUGUAAGUUGCUUACUUAAAUGCUACAUCCUUUGCCACAAAGUGUUAAAUGUAGCUUUU__AAAUGCACACAACCUUUAUUU .................(((((......(((.(((((((....(((......)))....))))))).)))......))).))........... (-11.39 = -11.70 + 0.31)

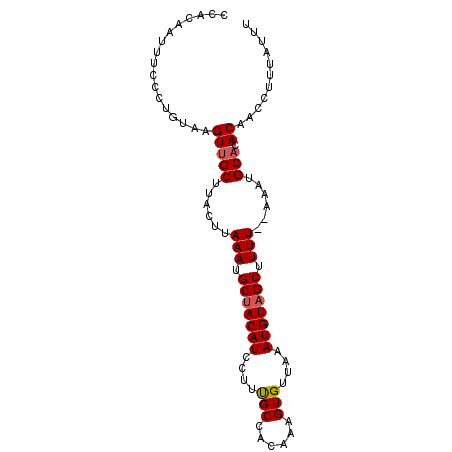
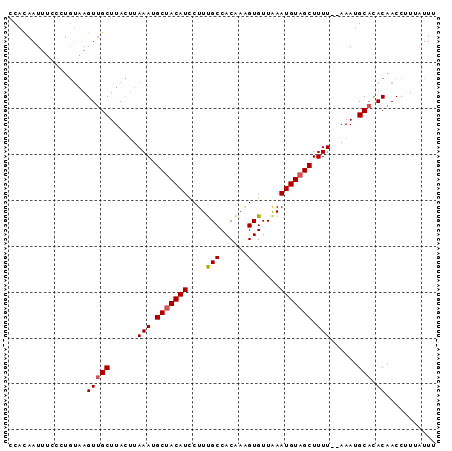
| Location | 19,710,336 – 19,710,456 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -17.06 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19710336 120 + 22407834 GUGCAUUUAUAAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUGUGGCAGCUUUCAUGCUGGUGCAUGCUAUCAAUCUGUCUCUGUA .(((.....((((.(((((((((..(((...))).....))))))))).)))).....)))...((((((((.(((((((((((..((.....)).)).))))).))))...)))))))) ( -33.00) >DroSim_CAF1 3695 116 + 1 GUGCAUUU--AAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUGUGGCAGCUUUCAU--UGGUGCAUGCCAUCAAUCUGUUUCUGUA .(((((((--(((.(((((((((..(((...))).....))))))))).)))))))..)))...(((((..(.(((((((((((......--....)).))))).))))...)..))))) ( -35.40) >DroEre_CAF1 3647 99 + 1 AAGCAUUU--AAAAGCUACAUUCAACACCACGUGGCAAAGGAUGUGGCAUUUAAAUAAGCGACU-----GCAAAUUAUGGCAGGUUUCA--------------AUCAAUCUAUUUCUGUU ..((((((--(((.(((((((((....((....))....))))))))).)))))))..))..((-----((........))))......--------------................. ( -24.00) >DroYak_CAF1 3718 116 + 1 AUGCAUUU--AAAAGCUACAUUUAACACUUUGUGGCGAAGAAUGUAGCAUUUAGGUAAGCAACUUACAGGGAAAUUAUGGCAGUUUUCAU--UGGUGCAUGCUAUCCAUCUAUUUCUGUU .(((((((--(((.(((((((((..(.((....)).)..))))))))).)))))))..)))....((((..(....((((((((......--....)).)))))).......)..)))). ( -26.30) >consensus AUGCAUUU__AAAAGCUACAUUUAACACUUUGUGGCAAAGGAUGUAGCAUUUAAGUAAGCAACUUACAGGGAAAUUAUGGCAGCUUUCAU__UGGUGCAUGCUAUCAAUCUAUUUCUGUA ..((......(((.(((((((((..(((...))).....))))))))).)))......)).....(((((((....((((((((............)).)))))).......))))))). (-17.06 = -18.62 + 1.56)

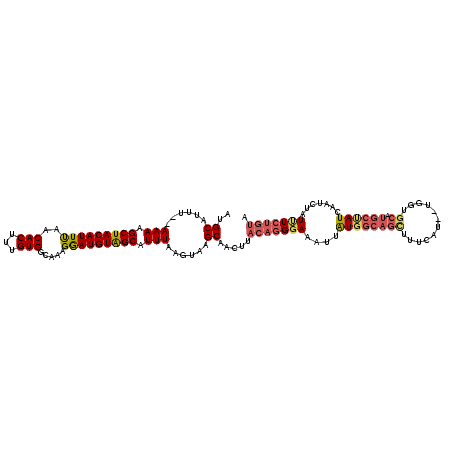
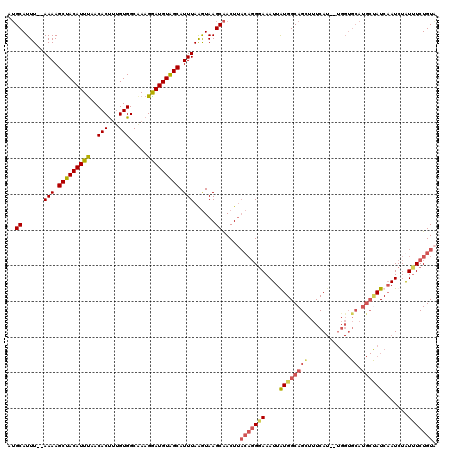
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:06 2006