| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,693,065 – 19,693,163 |
| Length | 98 |
| Max. P | 0.935566 |

| Location | 19,693,065 – 19,693,163 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -13.01 |
| Energy contribution | -16.15 |
| Covariance contribution | 3.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
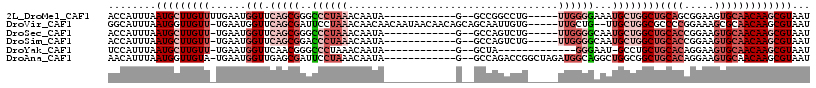
>2L_DroMel_CAF1 19693065 98 + 22407834 ACCAUUUAAUGCUUGUUUUGAAUGGUUCAGCGGGCCCUAAACAAUA------------G--GCCGGCCUG-----UUGGGGAAAUGCUGGCUGCAGCGGAAGUGCAACAAGCGUAAU ........(((((((((......(((.(((((..(((....(((((------------(--(....))))-----))))))...))))))))(((.(....)))))))))))))... ( -36.90) >DroVir_CAF1 117966 109 + 1 GGCAUUUAAUGGUUGUU-UGAAUGGUUCAGCGAUUCCUAAACAACAACAAUAACAACAGCAGCAAUUGUG-----UUGCUG--UUGCUGGCGCCCCGGAAAGCGCAACAAGCGUAAU (((........((((((-(((((.(.....).))))..)))))))........(((((((((((....))-----))))))--))).....))).......((((.....))))... ( -34.90) >DroSec_CAF1 114814 97 + 1 ACCAUUUAAUGCUUGUU-UGAAUGGUUCAGCGGGCCCUAAACAAUA------------G--GCCAGUCUG-----UUGGGGCAAUGCUGGCUGCACCGGAAGUGCAACAAGCGUAAU ........(((((((((-.....(((.(((((.((((....(((((------------(--(....))))-----)))))))..))))))))(((.(....)))))))))))))... ( -35.90) >DroSim_CAF1 94040 97 + 1 ACCAUUUAAUGCUUGUU-UGAAUGGUUCAGCGGACCCUAAACAAUA------------G--GCCAGUCUG-----UUGGGGCAAUGCUGGCUGCACCGGAAGUGCAACAAGCGUAAU (((((((((.......)-))))))))(((((((((((((.....))------------)--)...)))))-----))))....(((((.(.((((.(....))))).).)))))... ( -33.30) >DroYak_CAF1 89770 88 + 1 UCCAUUUAAUGCUUGUU-UGAAUGGUUCAACGGGCCCUAAACAAUA------------G--GCUA-------------GGGAAU-GCCUGCUGCACAGGAAGUGCAACAAGCGUAAU .((((((((.......)-))))))).........(((((..(....------------)--..))-------------))).((-((.((.(((((.....))))).)).))))... ( -26.00) >DroAna_CAF1 90941 102 + 1 AACAUUUAAUGGUUGUA-UGAAUGGUUGAGCGAUUCCUAAACAAUA------------G--GCCAGACCGGCUAGAUGGCAGGCUGGCGGCUGCACAGGAAGUGCAACAAGCGUAAU ........(((.((((.-.((((.(.....).))))..........------------.--(((((.((.(((....))).)))))))...(((((.....))))))))).)))... ( -29.40) >consensus ACCAUUUAAUGCUUGUU_UGAAUGGUUCAGCGGGCCCUAAACAAUA____________G__GCCAGUCUG_____UUGGGGAAAUGCUGGCUGCACCGGAAGUGCAACAAGCGUAAU ........(((((((((......(((.(((((..((((((...................................))))))...))))))))((((.....)))))))))))))... (-13.01 = -16.15 + 3.14)
| Location | 19,693,065 – 19,693,163 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -14.74 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
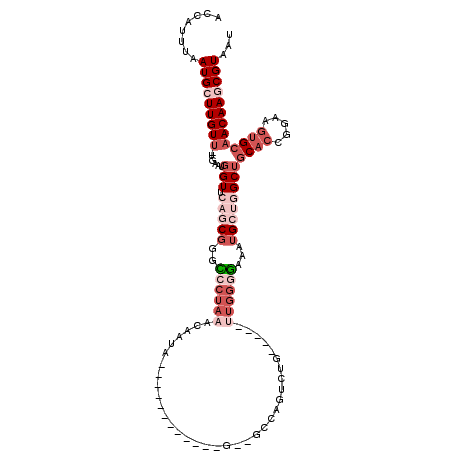
>2L_DroMel_CAF1 19693065 98 - 22407834 AUUACGCUUGUUGCACUUCCGCUGCAGCCAGCAUUUCCCCAA-----CAGGCCGGC--C------------UAUUGUUUAGGGCCCGCUGAACCAUUCAAAACAAGCAUUAAAUGGU .....(((.((((((.......)))))).)))..........-----..(((.(((--(------------(........))))).)))..((((((.((........)).)))))) ( -27.30) >DroVir_CAF1 117966 109 - 1 AUUACGCUUGUUGCGCUUUCCGGGGCGCCAGCAA--CAGCAA-----CACAAUUGCUGCUGUUGUUAUUGUUGUUGUUUAGGAAUCGCUGAACCAUUCA-AACAACCAUUAAAUGCC .....((.....((((((....)))))).(((((--(((((.-----((....)).))))))))))...((((((((((((......))))))......-))))))........)). ( -30.10) >DroSec_CAF1 114814 97 - 1 AUUACGCUUGUUGCACUUCCGGUGCAGCCAGCAUUGCCCCAA-----CAGACUGGC--C------------UAUUGUUUAGGGCCCGCUGAACCAUUCA-AACAAGCAUUAAAUGGU .....(((.(((((((.....))))))).)))..........-----(((.(.(((--(------------(........))))).)))).((((((.(-(.......)).)))))) ( -30.50) >DroSim_CAF1 94040 97 - 1 AUUACGCUUGUUGCACUUCCGGUGCAGCCAGCAUUGCCCCAA-----CAGACUGGC--C------------UAUUGUUUAGGGUCCGCUGAACCAUUCA-AACAAGCAUUAAAUGGU .....(((.(((((((.....))))))).)))..........-----(((.(.(((--(------------(........))))).)))).((((((.(-(.......)).)))))) ( -27.80) >DroYak_CAF1 89770 88 - 1 AUUACGCUUGUUGCACUUCCUGUGCAGCAGGC-AUUCCC-------------UAGC--C------------UAUUGUUUAGGGCCCGUUGAACCAUUCA-AACAAGCAUUAAAUGGA .....(((((((((((.....)))))))))))-...(((-------------(((.--(------------....).)))))).........(((((.(-(.......)).))))). ( -28.50) >DroAna_CAF1 90941 102 - 1 AUUACGCUUGUUGCACUUCCUGUGCAGCCGCCAGCCUGCCAUCUAGCCGGUCUGGC--C------------UAUUGUUUAGGAAUCGCUCAACCAUUCA-UACAACCAUUAAAUGUU .....(.(((..((..((((((.((((..(((((((.((......)).)).)))))--.------------..)))).))))))..)).))).).....-................. ( -26.60) >consensus AUUACGCUUGUUGCACUUCCGGUGCAGCCAGCAUUGCCCCAA_____CAGACUGGC__C____________UAUUGUUUAGGGCCCGCUGAACCAUUCA_AACAAGCAUUAAAUGGU .....(((.(((((((.....))))))).)))...........................................((((((......))))))........................ (-14.74 = -15.38 + 0.64)

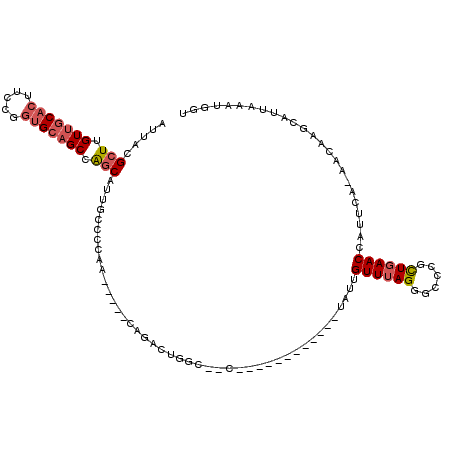
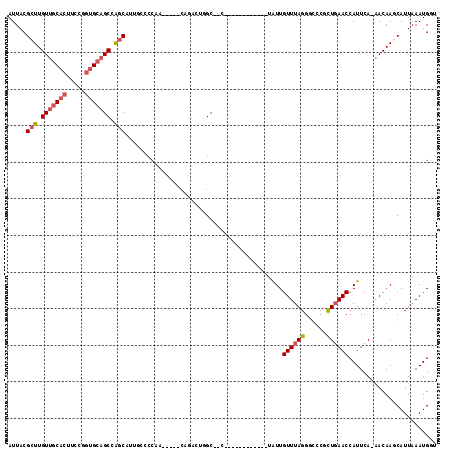
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:00 2006