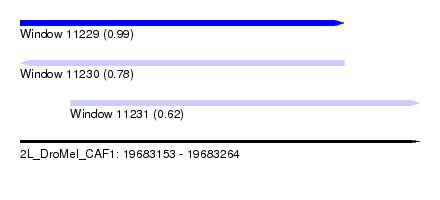
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,683,153 – 19,683,264 |
| Length | 111 |
| Max. P | 0.992640 |

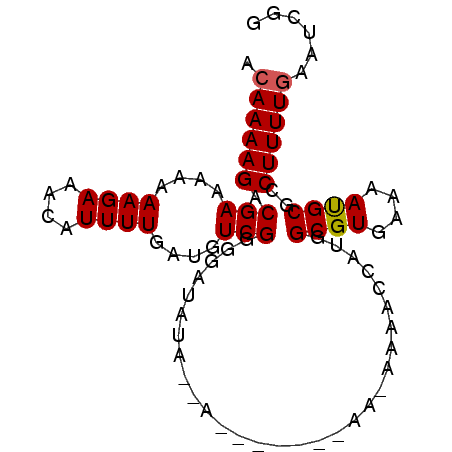
| Location | 19,683,153 – 19,683,243 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -15.15 |
| Consensus MFE | -12.74 |
| Energy contribution | -13.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19683153 90 + 22407834 ACAAAAGACGAAAAAAAGAAACAUUUUGAUGUCGGGUAUAUACAAAAAAAAAAAAAAAACCAUGGCGUGAAAAUGCGCCUUUUGAAUUGG .((((((.((...........((((((.(((((((((.....................))).)))))).)))))))).))))))...... ( -17.00) >DroSec_CAF1 104931 80 + 1 ACAAAAGACGAAAAAAAGAAACAUUUUGAUGUCGGGGAUACA--A-------AA-AAAAGCAUGGCGUGAAAAUGCGCCUUUUGAAUGGG .(((((((((...((((......))))..)))).........--.-------..-........(((((......))))))))))...... ( -14.00) >DroSim_CAF1 84230 81 + 1 ACAAAAGACGAAAAAAAGAAACAUUUUGAUGUCGGGGAUACA--A-------AAAAAAAGCAUGGCGUGAAAAUGCGCCUUUUGAAUUGG .(((((((((...((((......))))..)))).........--.-------...........(((((......))))))))))...... ( -14.00) >DroEre_CAF1 81111 79 + 1 ACAAAAGACGAAAAAAAGAAACAUUUUGAUGUCGGAGAUAUA--A-------AA--AAACCAUGGCGUGAAAACGCGCCUUUUGAAACGG .(((((((((...((((......))))..)))).........--.-------..--.......((((((....)))))))))))...... ( -15.60) >DroYak_CAF1 80455 79 + 1 ACAAAAGACGAAA-AAAGAAACAUUUUGAUGUCGGAUGAAUA--A-------AA-AAAAACAUGGCGUGAAAACGCGCCUUUUCAAACGG ......((((..(-(((......))))..))))(..((((..--.-------..-........((((((....))))))..))))..).. ( -15.16) >consensus ACAAAAGACGAAAAAAAGAAACAUUUUGAUGUCGGGGAUAUA__A_______AA_AAAACCAUGGCGUGAAAAUGCGCCUUUUGAAUCGG .(((((((((...((((......))))..))))..............................(((((......))))))))))...... (-12.74 = -13.14 + 0.40)

| Location | 19,683,153 – 19,683,243 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -12.06 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19683153 90 - 22407834 CCAAUUCAAAAGGCGCAUUUUCACGCCAUGGUUUUUUUUUUUUUUUUGUAUAUACCCGACAUCAAAAUGUUUCUUUUUUUCGUCUUUUGU ......(((((((((..............(((.....................))).(((((....))))).........))))))))). ( -10.90) >DroSec_CAF1 104931 80 - 1 CCCAUUCAAAAGGCGCAUUUUCACGCCAUGCUUUU-UU-------U--UGUAUCCCCGACAUCAAAAUGUUUCUUUUUUUCGUCUUUUGU ......(((((((((............((((....-..-------.--.))))....(((((....))))).........))))))))). ( -12.00) >DroSim_CAF1 84230 81 - 1 CCAAUUCAAAAGGCGCAUUUUCACGCCAUGCUUUUUUU-------U--UGUAUCCCCGACAUCAAAAUGUUUCUUUUUUUCGUCUUUUGU ......((((((((((((.........)))).......-------.--.........(((((....)))))..........)))))))). ( -12.00) >DroEre_CAF1 81111 79 - 1 CCGUUUCAAAAGGCGCGUUUUCACGCCAUGGUUU--UU-------U--UAUAUCUCCGACAUCAAAAUGUUUCUUUUUUUCGUCUUUUGU ......((((((((((((....))))........--..-------.--.........(((((....)))))..........)))))))). ( -13.80) >DroYak_CAF1 80455 79 - 1 CCGUUUGAAAAGGCGCGUUUUCACGCCAUGUUUUU-UU-------U--UAUUCAUCCGACAUCAAAAUGUUUCUUU-UUUCGUCUUUUGU .......(((((((((((....)))).........-..-------.--.........(((((....))))).....-....))))))).. ( -11.60) >consensus CCAAUUCAAAAGGCGCAUUUUCACGCCAUGCUUUU_UU_______U__UAUAUCCCCGACAUCAAAAUGUUUCUUUUUUUCGUCUUUUGU ......((((((((((........))...............................(((((....)))))..........)))))))). ( -9.94 = -10.14 + 0.20)

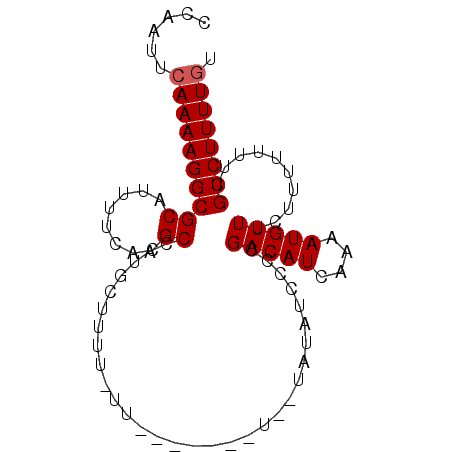
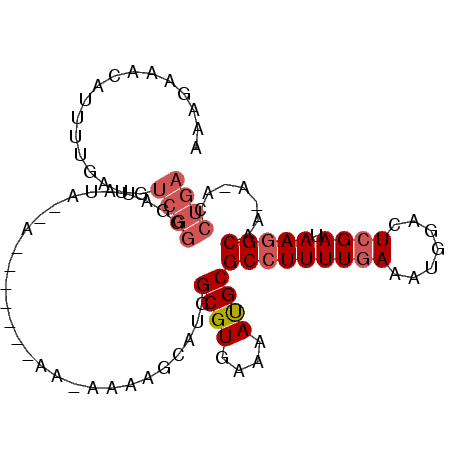
| Location | 19,683,167 – 19,683,264 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -11.62 |
| Energy contribution | -13.90 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19683167 97 + 22407834 AAAGAAACAUUUUGAUGUCGGGUAUAUACAAAAAAAAAAAAAAAACCAUGGCGUGAAAAUGCGCCUUUUGAAUUGGACUCGAUAAGGCAA-A-ACCUGA .......((((((.(((((((((.....................))).)))))).)))))).(((((((((.......)))).)))))..-.-...... ( -20.90) >DroSec_CAF1 104945 87 + 1 AAAGAAACAUUUUGAUGUCGGGGAUACA--A-------AA-AAAAGCAUGGCGUGAAAAUGCGCCUUUUGAAUGGGACUCGAUAAGGCAA-A-ACCUGA .................(((((......--.-------..-.........((((....))))(((((((((.......)))).)))))..-.-.))))) ( -19.80) >DroSim_CAF1 84244 88 + 1 AAAGAAACAUUUUGAUGUCGGGGAUACA--A-------AAAAAAAGCAUGGCGUGAAAAUGCGCCUUUUGAAUUGGACUCGAUAAGGCAA-A-ACCUGA .................(((((......--.-------............((((....))))(((((((((.......)))).)))))..-.-.))))) ( -19.80) >DroEre_CAF1 81125 86 + 1 AAAGAAACAUUUUGAUGUCGGAGAUAUA--A-------AA--AAACCAUGGCGUGAAAACGCGCCUUUUGAAACGGACUCGAUAAGGCAA-A-ACCUGA .................((((.......--.-------..--........((((....))))(((((((((.......)))).)))))..-.-..)))) ( -17.90) >DroYak_CAF1 80468 88 + 1 AAAGAAACAUUUUGAUGUCGGAUGAAUA--A-------AA-AAAAACAUGGCGUGAAAACGCGCCUUUUCAAACGGAGUCGAUAAGGCAAAA-ACCUGA ...........(((((..((..((((..--.-------..-........((((((....))))))..))))..))..)))))..(((.....-.))).. ( -19.26) >DroAna_CAF1 82060 78 + 1 AAGGAAACGCUUUGAA----------UG--G-------GC-AAAAGCACGGCGUGAAAAUGCGCCUUUUGAAAUGGACUCGAUAAAGCAA-ACACCUGA ..(....)((((((..----------((--(-------((-.....((.(((((......)))))...)).....).)))).))))))..-........ ( -18.00) >consensus AAAGAAACAUUUUGAUGUCGGGGAUAUA__A_______AA_AAAAGCAUGGCGUGAAAAUGCGCCUUUUGAAAUGGACUCGAUAAGGCAA_A_ACCUGA .................((((.............................((((....))))(((((((((.......)))).))))).......)))) (-11.62 = -13.90 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:55 2006