| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,643,595 – 19,643,699 |
| Length | 104 |
| Max. P | 0.703307 |

| Location | 19,643,595 – 19,643,699 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19643595 104 + 22407834 UUU-------UUUUUGGCAAGAUAUUGCUCGACAGAUGAGGAGACAGAGUCACGUUACUUGCAGGUGUUGCAAGUGUUGCAGGUGUUGCAGGUGCAAUCUAUCCAAAAAAA .((-------(((((((..((((..(((......(((..(....)...))).....((((((((...(((((.....)))))...)))))))))))))))..))))))))) ( -28.50) >DroSec_CAF1 86785 99 + 1 UU----------UUUGGCAAGAUAUUGCUCGACAGGUGAGGAGACAGAGUCACGUUACUUGCAGGUGUUGCAAGUGUUGCAGGUGUUGCAGGUGCAAUCUAUCCAAA--AA .(----------(((((..((((...((..(.(((((((.(.(((...))).).))))))))..))..((((..(((.((....)).)))..))))))))..)))))--). ( -28.90) >DroSim_CAF1 64900 99 + 1 UU----------UUUGGCAAGAUAUUGCUCGACAGGUGAGGAGACAGAGUCACGUUACUUGCAGGUGUUGCAAGUGUUGCAGGUGUUGCAGGUGCAAUCUAUCCAAA--AA .(----------(((((..((((...((..(.(((((((.(.(((...))).).))))))))..))..((((..(((.((....)).)))..))))))))..)))))--). ( -28.90) >DroEre_CAF1 62870 80 + 1 U-----------UUUGGCAAGAUAUUGCUCGACAGGUGAGGAGACAGAGUCACGUUACUUGCAGG------------------UGUUGCAGGUGCAAUCUAUCCAAA--AA (-----------(((((..((((...((..(.(((((((.(.(((...))).).))))))))..)------------------)..(((....)))))))..)))))--). ( -22.00) >DroYak_CAF1 62527 80 + 1 U-----------UUUGGCAAGAUAUUGCUCGACAGGUGAGGAGACAGAGUCACGUUACUUGCAGG------------------UGUUGCAGGUGCAAUCUAUCCAAA--AA (-----------(((((..((((...((..(.(((((((.(.(((...))).).))))))))..)------------------)..(((....)))))))..)))))--). ( -22.00) >DroAna_CAF1 64256 91 + 1 UCAGCUACAGCUUCAGUCAAGAUAUUGCUCGGCAGGUGAGGAGACAGAGUCACGUUACUUGCAGG------------------UGUUGCAGGUGCAAUCUAUCCAGG--GC ..(((....)))..............((((.((((((((.(.(((...))).).)))))))).((------------------.(((((....)))))....)).))--)) ( -25.60) >consensus UU__________UUUGGCAAGAUAUUGCUCGACAGGUGAGGAGACAGAGUCACGUUACUUGCAGG__________________UGUUGCAGGUGCAAUCUAUCCAAA__AA ............(((((..((((..(((.......(((((....)....))))...((((((((...(((((.....)))))...)))))))))))))))..))))).... (-18.51 = -19.22 + 0.71)

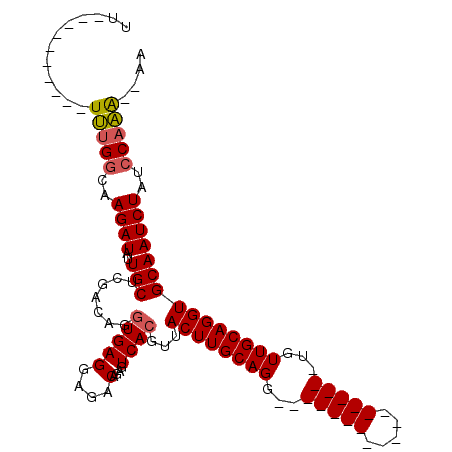
| Location | 19,643,595 – 19,643,699 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19643595 104 - 22407834 UUUUUUUGGAUAGAUUGCACCUGCAACACCUGCAACACUUGCAACACCUGCAAGUAACGUGACUCUGUCUCCUCAUCUGUCGAGCAAUAUCUUGCCAAAAA-------AAA ((((((((((((((...(((.((((.....))))..(((((((.....)))))))...)))..))))))..............((((....))))))))))-------)). ( -27.40) >DroSec_CAF1 86785 99 - 1 UU--UUUGGAUAGAUUGCACCUGCAACACCUGCAACACUUGCAACACCUGCAAGUAACGUGACUCUGUCUCCUCACCUGUCGAGCAAUAUCUUGCCAAA----------AA .(--((((((((((...(((.((((.....))))..(((((((.....)))))))...)))..))))))..............((((....))))))))----------). ( -24.90) >DroSim_CAF1 64900 99 - 1 UU--UUUGGAUAGAUUGCACCUGCAACACCUGCAACACUUGCAACACCUGCAAGUAACGUGACUCUGUCUCCUCACCUGUCGAGCAAUAUCUUGCCAAA----------AA .(--((((((((((...(((.((((.....))))..(((((((.....)))))))...)))..))))))..............((((....))))))))----------). ( -24.90) >DroEre_CAF1 62870 80 - 1 UU--UUUGGAUAGAUUGCACCUGCAACA------------------CCUGCAAGUAACGUGACUCUGUCUCCUCACCUGUCGAGCAAUAUCUUGCCAAA-----------A .(--(((((..(((((((((.((((...------------------..)))).))....((((..((......))...)))).)))..))))..)))))-----------) ( -17.20) >DroYak_CAF1 62527 80 - 1 UU--UUUGGAUAGAUUGCACCUGCAACA------------------CCUGCAAGUAACGUGACUCUGUCUCCUCACCUGUCGAGCAAUAUCUUGCCAAA-----------A .(--(((((..(((((((((.((((...------------------..)))).))....((((..((......))...)))).)))..))))..)))))-----------) ( -17.20) >DroAna_CAF1 64256 91 - 1 GC--CCUGGAUAGAUUGCACCUGCAACA------------------CCUGCAAGUAACGUGACUCUGUCUCCUCACCUGCCGAGCAAUAUCUUGACUGAAGCUGUAGCUGA ..--...((((((((..(((.((((...------------------..)))).))...)..).)))))))..(((.((((..(((....((......)).))))))).))) ( -18.10) >consensus UU__UUUGGAUAGAUUGCACCUGCAACA__________________CCUGCAAGUAACGUGACUCUGUCUCCUCACCUGUCGAGCAAUAUCUUGCCAAA__________AA .....((((..(((((((((.((((.....((((.....)))).....)))).))....((((..((......))...)))).)))..))))..))))............. (-12.98 = -13.70 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:49 2006