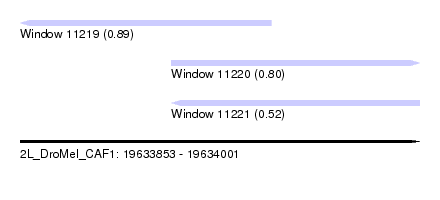
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,633,853 – 19,634,001 |
| Length | 148 |
| Max. P | 0.894344 |

| Location | 19,633,853 – 19,633,946 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19633853 93 - 22407834 AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAACCAAAAUCACUAAAGCCCAAGGAGAUUCCCAAAUUGAAGGAGAUUUCAGAAGGACU ...(.((((((((.((....)).)))))))).).................((((....(((((..((........))..)))))....)).)) ( -23.70) >DroSec_CAF1 77195 93 - 1 AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAACCAAAAUCACCAAAGCCCAAGGAGAUUCCCAAAUUGAAGGAGAUUUCAGAAGGACU ...(.((((((((.((....)).)))))))).).................((((....(((((..((........))..)))))....)).)) ( -23.70) >DroSim_CAF1 54593 93 - 1 AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAACCAAAAUCACCAAAGCCCAAGGAGAUUCCCAAAUUGAAGGAGAUUUCAGAAGGACU ...(.((((((((.((....)).)))))))).).................((((....(((((..((........))..)))))....)).)) ( -23.70) >DroEre_CAF1 53244 93 - 1 AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAGAAACCAAAAUCACCAAAGCCCAAGGAUAUCCCCAAAUUGAAGGAGAUUUCAGAAAGGUU ..((.((((((((.((....)).)))))))).))................((((....((.(((.((........)).))).)).....)))) ( -25.50) >DroYak_CAF1 53077 93 - 1 AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAGAAACCAAAAGCAUCAAAGCCCAAGGAUAUUCCCAAAUUGAAGGAGAUUUCAGAAAGAUU ..((.((((((((.((....)).)))))))).))...((....((......))....))...........((((((....))))))....... ( -22.40) >DroAna_CAF1 55063 84 - 1 AACUGUCCGGGCACUUGCCAAAUUGCCAGGAGAAAAACCA---------AAGCCUAAAUAGAUCCCCAAAUUGAAGGAGAUUUUUUAAGGUUC ...(.(((.((((.((....)).)))).))).).......---------.(((((.((.(((((.((........)).))))).)).))))). ( -20.30) >consensus AACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAACCAAAAUCACCAAAGCCCAAGGAGAUUCCCAAAUUGAAGGAGAUUUCAGAAGGACU ...(.((((((((.((....)).)))))))).).........................((((((.((........)).))))))......... (-19.09 = -19.53 + 0.45)

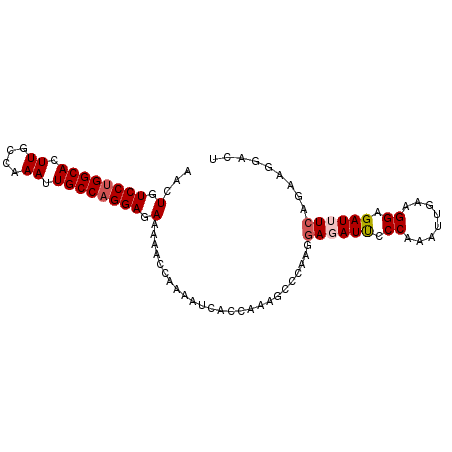

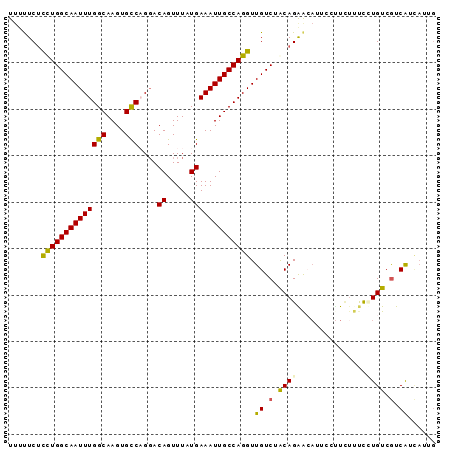
| Location | 19,633,909 – 19,634,001 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19633909 92 + 22407834 UUUUUCUCCUGGCAAUUUGGCAAGUGCCAGGACAGUUUAUGAAAUUGCCAGGUUGUCUGCAGGACAUGCCCUCUUUCCUGUCGUCAUCAUUG .......((((((((((((((....)))....((.....))))))))))))).((.(.((((((...........)))))).).))...... ( -28.60) >DroSim_CAF1 54649 92 + 1 UUUUUCUCCUGGCAAUUUGGCAAGUGCCAGGACAGUUUAUGAAAUUGCCAGGUUGUCUGCAGGACAUGCCUUCUUUCCUGUCGUCAUCAUUG .......((((((((((((((....)))....((.....))))))))))))).((.(.((((((...........)))))).).))...... ( -28.60) >DroEre_CAF1 53300 92 + 1 UUUCUCUCCUGGCAAUUUGGCAAGUGCCAGGACAGUUUAUGAAAUUGCCAGGUUGUCUACAGAACAUUCCUUCUUUCUUGUCGUCAUCAUUG .......((((((((((((((....)))....((.....))))))))))))).((.(.((((((..........))).))).).))...... ( -22.20) >DroWil_CAF1 84170 82 + 1 UCCUUCAUUUGGCAAUUUGGCAAGUGUCUGCACAGUUUAUGAAAUUGCCAGGUUGUCUACAGAAA------UAUUCCAUGUCGUCGUU---- ...(((....(((((((((((((...((............))..)))))))))))))....))).------.................---- ( -19.60) >DroYak_CAF1 53133 92 + 1 UUUCUCUCCUGGCAAUUUGGCAAGUGCCAGGACAGUUUAUGAAAUUGCCAGGUUGUCUACAGAGCAUUCCUUCUCUCUUGUCGUCAUCAUUG .......((((((((((((((....)))....((.....))))))))))))).((.(.((((((..........))).))).).))...... ( -23.70) >DroAna_CAF1 55110 90 + 1 UUUUUCUCCUGGCAAUUUGGCAAGUGCCCGGACAGUUUAUGAAAUUGCCAGGUUGUCCGCAGAAAAUUCCUUU--UUGUGUCAUCAUGAUUG .......((((((((((((((....)))....((.....))))))))))))).....((((((((.....)))--)))))............ ( -25.20) >consensus UUUUUCUCCUGGCAAUUUGGCAAGUGCCAGGACAGUUUAUGAAAUUGCCAGGUUGUCUACAGAACAUUCCUUCUUUCCUGUCGUCAUCAUUG .......((((((((((((((....)))....((.....))))))))))))).((.(.((((...............)))).).))...... (-20.59 = -20.18 + -0.41)
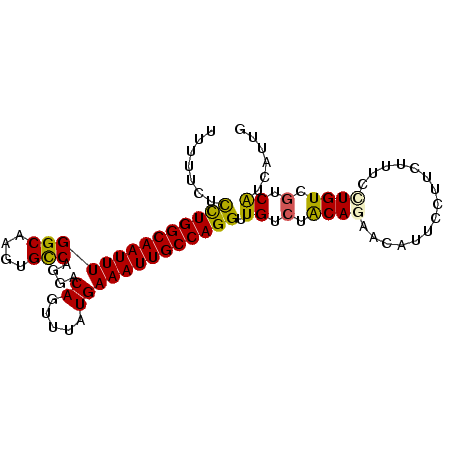
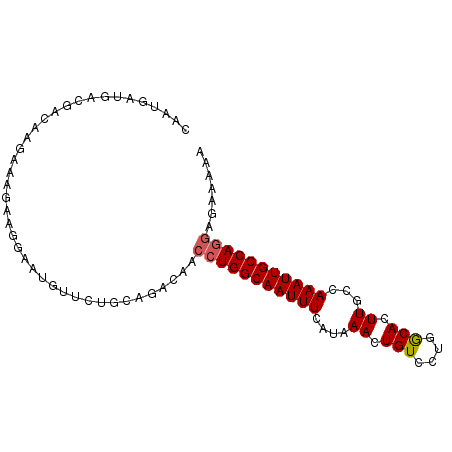
| Location | 19,633,909 – 19,634,001 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -21.72 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.13 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19633909 92 - 22407834 CAAUGAUGACGACAGGAAAGAGGGCAUGUCCUGCAGACAACCUGGCAAUUUCAUAAACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAA ......((.(..(((((..(....)...)))))..).)).((((((((((.............((((....))))))))))))))....... ( -25.61) >DroSim_CAF1 54649 92 - 1 CAAUGAUGACGACAGGAAAGAAGGCAUGUCCUGCAGACAACCUGGCAAUUUCAUAAACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAA ......((.(..(((((..(....)...)))))..).)).((((((((((.............((((....))))))))))))))....... ( -25.61) >DroEre_CAF1 53300 92 - 1 CAAUGAUGACGACAAGAAAGAAGGAAUGUUCUGUAGACAACCUGGCAAUUUCAUAAACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAGAAA ...............((((..(((..((((.....)))).))).....)))).....((.((((((((.((....)).)))))))).))... ( -23.40) >DroWil_CAF1 84170 82 - 1 ----AACGACGACAUGGAAUA------UUUCUGUAGACAACCUGGCAAUUUCAUAAACUGUGCAGACACUUGCCAAAUUGCCAAAUGAAGGA ----.........(((((...------..)))))........((((((((........((.((((....))))))))))))))......... ( -11.90) >DroYak_CAF1 53133 92 - 1 CAAUGAUGACGACAAGAGAGAAGGAAUGCUCUGUAGACAACCUGGCAAUUUCAUAAACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAGAAA ......................((((((((..((.....))..))).))))).....((.((((((((.((....)).)))))))).))... ( -23.60) >DroAna_CAF1 55110 90 - 1 CAAUCAUGAUGACACAA--AAAGGAAUUUUCUGCGGACAACCUGGCAAUUUCAUAAACUGUCCGGGCACUUGCCAAAUUGCCAGGAGAAAAA .................--.......((((((........(((((((((((......(.....)(((....)))))))))))))))))))). ( -20.20) >consensus CAAUGAUGACGACAAGAAAGAAGGAAUGUUCUGCAGACAACCUGGCAAUUUCAUAAACUGUCCUGGCACUUGCCAAAUUGCCAGGAGAAAAA ........................................(((((((((((....((.(((....))).))...)))))))))))....... (-16.94 = -17.13 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:46 2006