
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,628,443 – 19,628,539 |
| Length | 96 |
| Max. P | 0.877944 |

| Location | 19,628,443 – 19,628,539 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -14.36 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
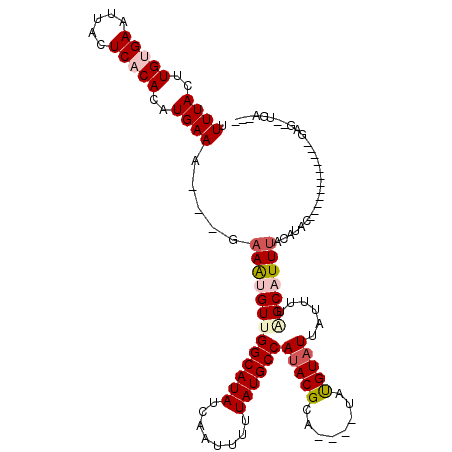
>2L_DroMel_CAF1 19628443 96 + 22407834 CUUUACUUGUGAAUUACUCACACAUGAAU---GAAAUGUUGGCAUAUCAAUUUUUAUGCCACACGCA----UACGUAUUAUUUUAGCAUUUACAUAC------------GAG--UGA--- ..((((((((((.....))))....(.((---((((((((((((((........))))))..(((..----..)))........))))))).))).)------------)))--)))--- ( -22.10) >DroPse_CAF1 68152 110 + 1 UUUUACUUGCGAAUUACUCACACAUGAAA---GAAAUGUUGGCAUAUCAAUUUUUAUGCCAUACGCA----UAUGUAUUAUUUCUGCAUUUACAUACUCAUACCCCUACGAAUAUGA--- ...(((.((((..........((((....---...))))(((((((........)))))))..))))----...)))...............((((.((..........)).)))).--- ( -18.40) >DroGri_CAF1 63479 97 + 1 UUUUACUUGUGAAUUACUCACACAUGAAA-----AACGUUGGCAUAUCAAUUUUUAUGCCAUACGCA----CACGUAUUAUUUUAGCAUUUACAUAU------------GCG--GGCACC (((((..(((((.....)))))..)))))-----...(((((((((........))))))).))((.----..(((((.................))------------)))--.))... ( -21.63) >DroWil_CAF1 76442 103 + 1 UUUUACUUGUGAAUUACUCACACAUGAAAAAAAAAAUGUUGGCAUAUCAAUUUUUAUGCCAUACGCACAUGUAUGUAUUAUUUUAGCAUUUACAUAU------------GAC--UUU--- (((((..(((((.....)))))..)))))........(((((((((........))))))).))......(((((((.............)))))))------------...--...--- ( -19.82) >DroMoj_CAF1 78859 95 + 1 AUUUACUUGUGAAUUACUCACACAUGAAA----AAGCGUUGGCAUAUCAAUUUUUAUGCCAUACGCA----CAUGUAUUAUUUUGGCAUUUACAUAU------------GAG--GGC--- .((((..(((((.....)))))..)))).----..(((((((((((........))))))).)))).----.(((((.............)))))..------------...--...--- ( -22.62) >DroPer_CAF1 71002 110 + 1 UUUUACUUGCGAAUUACUCACACAUGAAA---GAAAUGUUGGCAUAUCAAUUUUUAUGCCAUACGCA----UAUGUAUUAUUUCUGCAUUUACAUACUCAUACCCCUACGAAUAUGA--- ...(((.((((..........((((....---...))))(((((((........)))))))..))))----...)))...............((((.((..........)).)))).--- ( -18.40) >consensus UUUUACUUGUGAAUUACUCACACAUGAAA___GAAAUGUUGGCAUAUCAAUUUUUAUGCCAUACGCA____UAUGUAUUAUUUUAGCAUUUACAUAC____________GAG__UGA___ .((((..(((((.....)))))..)))).....(((((((((((((........))))))(((((........)))))......)))))))............................. (-14.36 = -15.25 + 0.89)

| Location | 19,628,443 – 19,628,539 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
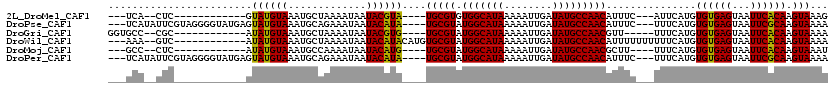
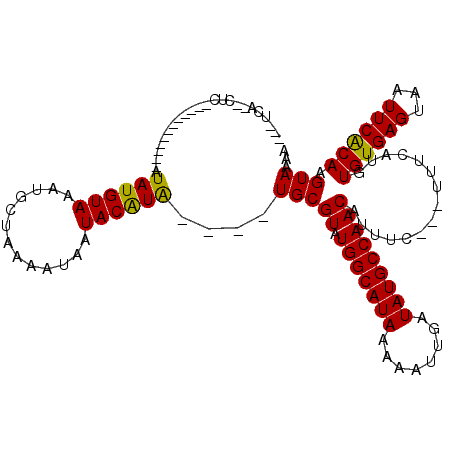
>2L_DroMel_CAF1 19628443 96 - 22407834 ---UCA--CUC------------GUAUGUAAAUGCUAAAAUAAUACGUA----UGCGUGUGGCAUAAAAAUUGAUAUGCCAACAUUUC---AUUCAUGUGUGAGUAAUUCACAAGUAAAG ---.((--(.(------------(((((((.............))))))----)).)))(((((((........))))))).......---.......((((((...))))))....... ( -21.12) >DroPse_CAF1 68152 110 - 1 ---UCAUAUUCGUAGGGGUAUGAGUAUGUAAAUGCAGAAAUAAUACAUA----UGCGUAUGGCAUAAAAAUUGAUAUGCCAACAUUUC---UUUCAUGUGUGAGUAAUUCGCAAGUAAAA ---.((((((((((....))))))))))....(((.(((.((.((((((----((.((.(((((((........))))))))).....---...))))))))..)).))))))....... ( -25.60) >DroGri_CAF1 63479 97 - 1 GGUGCC--CGC------------AUAUGUAAAUGCUAAAAUAAUACGUG----UGCGUAUGGCAUAAAAAUUGAUAUGCCAACGUU-----UUUCAUGUGUGAGUAAUUCACAAGUAAAA ..(((.--(((------------(((((.(((((........(((((..----..)))))((((((........))))))..))))-----)..)))))))).))).............. ( -25.90) >DroWil_CAF1 76442 103 - 1 ---AAA--GUC------------AUAUGUAAAUGCUAAAAUAAUACAUACAUGUGCGUAUGGCAUAAAAAUUGAUAUGCCAACAUUUUUUUUUUCAUGUGUGAGUAAUUCACAAGUAAAA ---...--(.(------------(((((((.................)))))))))((.(((((((........)))))))))...............((((((...))))))....... ( -18.93) >DroMoj_CAF1 78859 95 - 1 ---GCC--CUC------------AUAUGUAAAUGCCAAAAUAAUACAUG----UGCGUAUGGCAUAAAAAUUGAUAUGCCAACGCUU----UUUCAUGUGUGAGUAAUUCACAAGUAAAU ---...--...------------.........(((.........(((((----.((((.(((((((........)))))))))))..----...)))))(((((...)))))..)))... ( -21.70) >DroPer_CAF1 71002 110 - 1 ---UCAUAUUCGUAGGGGUAUGAGUAUGUAAAUGCAGAAAUAAUACAUA----UGCGUAUGGCAUAAAAAUUGAUAUGCCAACAUUUC---UUUCAUGUGUGAGUAAUUCGCAAGUAAAA ---.((((((((((....))))))))))....(((.(((.((.((((((----((.((.(((((((........))))))))).....---...))))))))..)).))))))....... ( -25.60) >consensus ___UCA__CUC____________AUAUGUAAAUGCUAAAAUAAUACAUA____UGCGUAUGGCAUAAAAAUUGAUAUGCCAACAUUUC___UUUCAUGUGUGAGUAAUUCACAAGUAAAA ........................((((((.............))))))....(((((.(((((((........)))))))))...............((((((...)))))).)))... (-15.78 = -15.12 + -0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:42 2006