| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,023,766 – 2,023,911 |
| Length | 145 |
| Max. P | 0.918767 |

| Location | 2,023,766 – 2,023,877 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
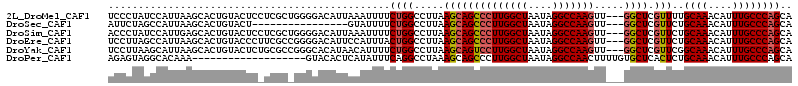
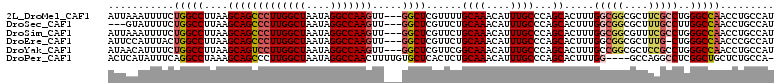
>2L_DroMel_CAF1 2023766 111 + 22407834 UCCCUAUCCAUUAAGCACUGUACUCCUCGCUGGGGACAUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUUUGCAAACAUUUGCCCAGCA ............................((((((.....(((((.((...((...(((((((((((((((....)))))))..---)))).)))).)).)).))))))))))). ( -30.30) >DroSec_CAF1 41131 95 + 1 AUUCUAGCCAUUAAGCACUGUACU----------------GUAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCA ......((((..(((.((......----------------)).)))..)))).....(.(((((((((((....)))))))..---)))))(((..((((....))))..))). ( -26.20) >DroSim_CAF1 39647 111 + 1 ACCCUAUCCAUUGAGCACUGUACUCCUCGCUGGGGACAUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCA ............(((.......)))...(((((((((.............((.....))(((((((((((....)))))))..---)))).)))).((((....))))))))). ( -31.10) >DroEre_CAF1 39793 111 + 1 UCCUUAGCCAUUAAGCACUGUACCCUUCGCCGGGGACAUUCCAUUUACUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCA ..((((((((.((((...(((.((((.....))))))).....)))).))))..)))).(((((((((((....)))))))..---)))).(((..((((....))))..))). ( -31.80) >DroYak_CAF1 43100 111 + 1 UCCUUAAGCAUUAAGCACUGUACUCUGCGCCGGGCACAUAACAUUUUCUGGCCUUAAGCAGUCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCGGCAAACAUUUGCCCAGCA ..((((((......(((........)))((((((............)))))))))))).(((((((((((....)))))))..---)))).(((.(((((....))))).))). ( -32.80) >DroPer_CAF1 51335 95 + 1 AGAGUAGGCACAAA-------------------GUACACUCAUAUUUCAGGCCUAAAGCAGCCCUUGGCUAAUAGGCCAACUUUUGUGCUCACUCUGCAAACAUUUGCCCAGCA (((((..(((((((-------------------(......)........((((((....((((...))))..))))))....)))))))..)))))((((....))))...... ( -31.10) >consensus ACCCUAGCCAUUAAGCACUGUACUC_UCGC_GGGGACAUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU___GGCUCGUUCUGCAAACAUUUGCCCAGCA ...............................................((((.....((((((((((((((....))))))).....)))).)))..((((....)))))))).. (-17.84 = -18.23 + 0.39)
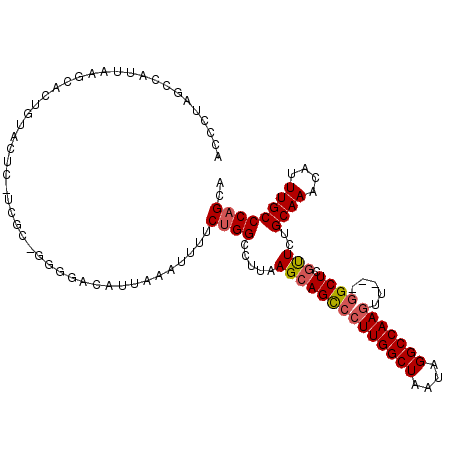
| Location | 2,023,803 – 2,023,911 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.61 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -26.75 |
| Energy contribution | -27.53 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.546167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
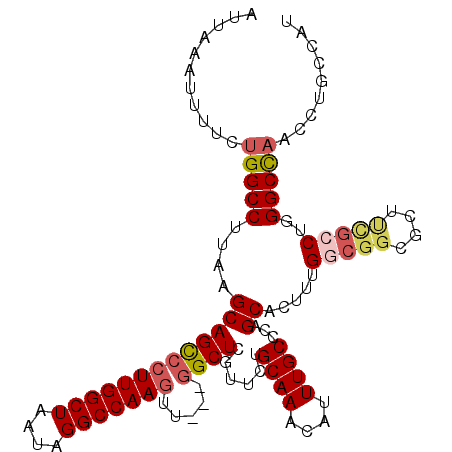
>2L_DroMel_CAF1 2023803 108 + 22407834 AUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUUUGCAAACAUUUGCCCAGCACUUUGGCGGCGCUUCGCCUGGGCCAACCUGCCAU ...........((((((.....((((...))))...)))))).(((---((((((((..((((....))))..))).....(((((....))))).))))))))....... ( -37.80) >DroSec_CAF1 41155 105 + 1 ---GUAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCACUUUGGCGGCGCUUUGCCUUGGCCAACCUGCCAU ---........(((((....(((((((((((((....)))))))..---)))).(((..((((....))))..)))......))(((.....)))..)))))......... ( -34.00) >DroSim_CAF1 39684 108 + 1 AUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCACUUUGGCGGCGUUUCGCCUGGGCCAACCUGCCAU ...........((((((.....((((...))))...)))))).(((---((((((((..((((....))))..))).....(((((....))))).))))))))....... ( -37.80) >DroEre_CAF1 39830 107 + 1 AUUCCAUUUACUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCUGCAAACAUUUGCCCAGCACUUUGGCGGCGCUUUG-CUGGGCCAACCCGCCAU ...........((((....((((((((((((((....)))))))..---)))).)))............((((((((....(((...))).))-))))))......)))). ( -37.90) >DroYak_CAF1 43137 108 + 1 AUAACAUUUUCUGGCCUUAAGCAGUCCUUGGCUAAUAGGCCAAGUU---GGCUCGUUCGGCAAACAUUUGCCCAGCACUUUGCCGGCGCUCCGCCUGGGCCAACCUGCCAU ...........((((((..(((........)))...)))))).(((---((((((((.(((((....))))).)))........((((...)))).))))))))....... ( -39.80) >DroPer_CAF1 51353 106 + 1 ACUCAUAUUUCAGGCCUAAAGCAGCCCUUGGCUAAUAGGCCAACUUUUGUGCUCACUCUGCAAACAUUUGCCCAGCACUUUGG----GCCAGGCCUCGGCUGCUCUGCCA- ............(((....(((((((...((((....((((.......(((((......((((....))))..)))))....)----))).))))..)))))))..))).- ( -38.10) >consensus AUUAAAUUUUCUGGCCUUAAGCAGCCCUUGGCUAAUAGGCCAAGUU___GGCUCGUUCUGCAAACAUUUGCCCAGCACUUUGGCGGCGCUUCGCCUGGGCCAACCUGCCAU ...........(((((....(((((((((((((....))))))).....))))......((((....))))...)).....(((((....)))))..)))))......... (-26.75 = -27.53 + 0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:21 2006