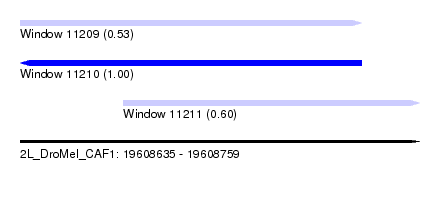
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,608,635 – 19,608,759 |
| Length | 124 |
| Max. P | 0.998517 |

| Location | 19,608,635 – 19,608,741 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
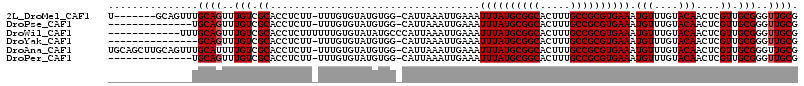
>2L_DroMel_CAF1 19608635 106 + 22407834 U-------GCAGUUUGCAGUUUGUCGCACCUCUU-UUUGUGUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG .-------((((..(((((..(((((((...(..-.....)..)))))-))...........((((((((((.....))))))))))................)))))..)))). ( -33.60) >DroPse_CAF1 41916 99 + 1 --------------UGCAGUUUGUCGCACCUCUU-UUUGUGUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG --------------(((((..(((((((...(..-.....)..)))))-))...........((((((((((.....))))))))))....)))))(((((((...))))))).. ( -29.80) >DroWil_CAF1 51583 103 + 1 ------------UUUGCAGUUUGUCGCACCUCUUUUUUGUGUAUAUGCCCAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG ------------...((((..(((.((....(......)((((((.((.((......))...((((((((((.....))))))))))..)).))))))....)).)))..)))). ( -28.30) >DroYak_CAF1 29405 98 + 1 ---------------GCAGUUUGUCGCACCUCUU-UUUGUGUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG ---------------.((((((((((((...(..-.....)..)))))-))...)))))...((((((((((.....)))))))))).........(((((((...))))))).. ( -29.70) >DroAna_CAF1 29513 113 + 1 UGCAGCUUGCAGUUUGCAUUUUGUCGCACCUCUU-UUUGUGUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG .(((((((((((((((((...(((((((...(..-.....)..)))))-))...........((((((((((.....)))))))))).....)))).)))....)))))))))). ( -38.70) >DroPer_CAF1 43533 99 + 1 --------------UGCAGUUUGUCGCACCUCUU-UUUGUGUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG --------------(((((..(((((((...(..-.....)..)))))-))...........((((((((((.....))))))))))....)))))(((((((...))))))).. ( -29.80) >consensus ______________UGCAGUUUGUCGCACCUCUU_UUUGUGUAUGUGG_CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCG ...............((((..(((.((...................................((((((((((.....)))))))))).(((....)))....)).)))..)))). (-25.38 = -25.55 + 0.17)
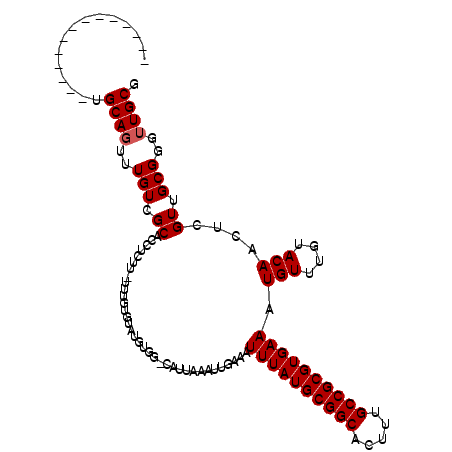
| Location | 19,608,635 – 19,608,741 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.31 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19608635 106 - 22407834 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG-CCACAUACACAAA-AAGAGGUGCGACAAACUGCAAACUGC-------A .(((....(((....(((((((...........(((((.....))))).(((((....)))))...-.............-.....)))))))....)))....)))-------. ( -24.90) >DroPse_CAF1 41916 99 - 1 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG-CCACAUACACAAA-AAGAGGUGCGACAAACUGCA-------------- ........(((....(((((((...........(((((.....))))).(((((....)))))...-.............-.....)))))))....))).-------------- ( -22.40) >DroWil_CAF1 51583 103 - 1 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUGGGCAUAUACACAAAAAAGAGGUGCGACAAACUGCAAA------------ ........(((....(((((((...........(((((.....)))))......(((..(((..((........))..)))..))))))))))....)))...------------ ( -22.60) >DroYak_CAF1 29405 98 - 1 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG-CCACAUACACAAA-AAGAGGUGCGACAAACUGC--------------- .((.....)).....(((((((...........(((((.....))))).(((((....)))))...-.............-.....))))))).......--------------- ( -21.10) >DroAna_CAF1 29513 113 - 1 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG-CCACAUACACAAA-AAGAGGUGCGACAAAAUGCAAACUGCAAGCUGCA .(((....((.....(((((((...........(((((.....))))).(((((....)))))...-.............-.....)))))))....(((.....))).))))). ( -26.20) >DroPer_CAF1 43533 99 - 1 CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG-CCACAUACACAAA-AAGAGGUGCGACAAACUGCA-------------- ........(((....(((((((...........(((((.....))))).(((((....)))))...-.............-.....)))))))....))).-------------- ( -22.40) >consensus CGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG_CCACAUACACAAA_AAGAGGUGCGACAAACUGCA______________ ........(((....(((((((...........(((((.....))))).(((((....))))).......................)))))))....)))............... (-22.12 = -22.12 + -0.00)


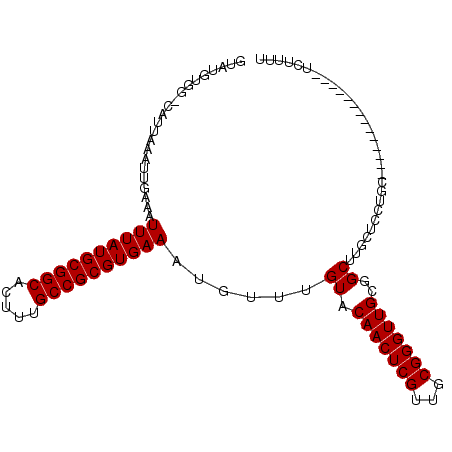
| Location | 19,608,667 – 19,608,759 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19608667 92 + 22407834 GUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUUGCUGCUGC--------------UGUUUU ....(..(-((...........((((((((((.....))))))))))..((..((.(((((((...)))))))..))..)))))..)--------------...... ( -31.00) >DroPse_CAF1 41941 92 + 1 GUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUCUUUCAUUC--------------UCUUUU ....((.(-(............((((((((((.....))))))))))..........((((((...)))))))).))..........--------------...... ( -24.40) >DroSim_CAF1 29393 92 + 1 GUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUUGCUGCUGC--------------UGUUUU ....(..(-((...........((((((((((.....))))))))))..((..((.(((((((...)))))))..))..)))))..)--------------...... ( -31.00) >DroWil_CAF1 51611 107 + 1 GUAUAUGCCCAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUUUCUCUCACACUCACUCUCAGACUCUAUC ......(((.............((((((((((.....)))))))))).........(((((((...))))))).))).............................. ( -26.10) >DroAna_CAF1 29552 88 + 1 GUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU-GCCUCUG-----------------UGUU ....(.((-((...........((((((((((.....))))))))))......((.(((((((...)))))))..)))-))).)..-----------------.... ( -28.00) >DroPer_CAF1 43558 92 + 1 GUAUGUGG-CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUCUUUCAUUC--------------UCUUUU ....((.(-(............((((((((((.....))))))))))..........((((((...)))))))).))..........--------------...... ( -24.40) >consensus GUAUGUGG_CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUUGCUCCUGC______________UCUUUU ......................((((((((((.....))))))))))......((.(((((((...)))))))..)).............................. (-22.10 = -22.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:36 2006