| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,575,958 – 19,576,065 |
| Length | 107 |
| Max. P | 0.714684 |

| Location | 19,575,958 – 19,576,065 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.81 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -13.57 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
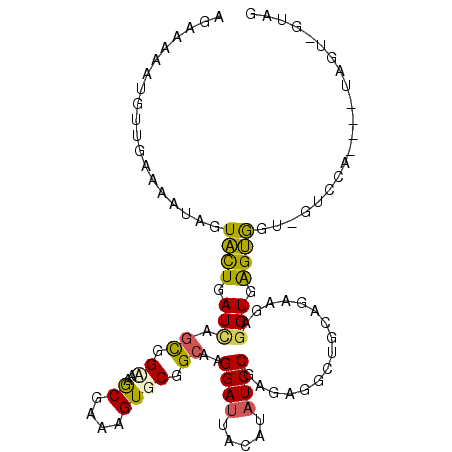
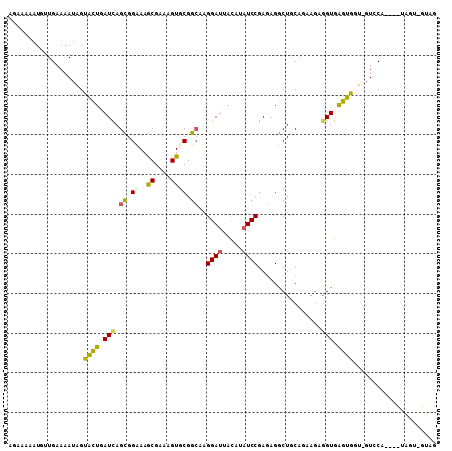
>2L_DroMel_CAF1 19575958 107 - 22407834 AGAAAAAUGAUGAAAAUUGUACUCAUCAGCGGCAAGCGAAAGUGCGGCAAGGAUUACAUAUCCGAGAGGCUGCAGCGGAGGUGGGUGCA-GUGCAUAGCUAGUCGUAG .((......(((...((((((((((((..(.((..((....))(((((..((((.....)))).....))))).)).).))))))))))-)).)))......)).... ( -37.90) >DroPse_CAF1 530 93 - 1 AGAAAAAUGUUGAAAAUAGUCUUAAUUAGUGGUAAACGCAAGUGCGGCAAGGAUUAUAUAUCCGAGAGACUACAAAAGCGGUAAGGGGU-GUAC-------------- .......((((.....(((((((.....((.((..((....)))).))..((((.....))))..)))))))....)))).........-....-------------- ( -19.00) >DroSim_CAF1 538 107 - 1 ACAAUAAUGCUGAAAAUUGUACUGAUCAGCGGAAAGCGAAAGUGCGGCAAGGAUUACAUAUCCGAGAGGCUGCAGCGAAGGUGGGUGCA-GUCCAAUGCUAGUGGUAG .((.....((.....((((((((.(((.((.....((....))(((((..((((.....)))).....))))).))...))).))))))-)).....))...)).... ( -32.00) >DroYak_CAF1 790 107 - 1 CGAAAAAUGUUGAAAAUUGUACUGAUCAGCGGCAAGCGAAAGUGCGGCAAGGAUUACAUAUCCGAGAGGCUGCAGCGGAGGUGGGUGGU-CUCCAUUAUGGGUCGUAG ...................(((.((((.((.((..((....)))).))..((((.....))))(.(((((..(.((....))..)..))-))))......))))))). ( -29.10) >DroAna_CAF1 834 107 - 1 AGA-AGAUGAUGAAAAUAGUACUGAUCAGUGGAAAGCGCAAGUGCGGAAAGGAUUACAUCUCCGAAAGGCUCCAUAAGACGUGAGUAUUUGUCCUGAAGUAUCAUUAC ...-.(((((((....(((.((......(((((...(((....)))...............((....)).)))))...((....))....)).)))...))))))).. ( -22.10) >DroPer_CAF1 1236 93 - 1 AGAAAAAUGUUGAAAAUAGUCUUAAUUAGUGGUAAACGCAAGUGCGGCAAGGAUUAUAUAUCCGAGAGACUUCAAAAGCGGUAAGGGGU-GUAC-------------- .......((((......((((((.....((.((..((....)))).))..((((.....))))..)))))).....)))).........-....-------------- ( -18.20) >consensus AGAAAAAUGUUGAAAAUAGUACUGAUCAGCGGAAAGCGAAAGUGCGGCAAGGAUUACAUAUCCGAGAGGCUGCAGAAGAGGUGAGUGGU_GUCCA____UAGU_GUAG ...................((((.(((.((.((..((....)))).))..((((.....))))................))).))))..................... (-13.57 = -12.60 + -0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:28 2006