
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,021,107 – 2,021,212 |
| Length | 105 |
| Max. P | 0.558467 |

| Location | 2,021,107 – 2,021,212 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -42.87 |
| Consensus MFE | -27.54 |
| Energy contribution | -28.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2021107 105 + 22407834 UGG-UUAGGGGCGGGUGGCAUGCCCCCGUGUGCGAGUGAGCGGCGAACAUCUGUUUGCCUUCAAAGUUGUGGCGUGCGGCUUGUGCGGGCUGCCCGAAUGCAAAGG ...-....((((((.(.(((((...(((..(((.(.((((.(((((((....)))))))))))......).)))..)))..))))).).))))))........... ( -42.90) >DroSec_CAF1 38564 104 + 1 UGG-UUAGGGGCGGGUGGCAUGCC-CCGUGUGCGAGUGAGCGGCGAACAUCUGUUUGCCUUCAAAGUUGCGGCGUGCGGCUUGUGCGGGCUGCCCGCAUGCAAAGG ...-...((((((.......))))-))(((((((..((((.(((((((....))))))))))).....(((((.((((.....)))).))))).)))))))..... ( -47.50) >DroSim_CAF1 37025 105 + 1 GGG-UUAGGGGCGGGUGGCAUGCCCCCGUGUGCGGGAGAGCGGCGAACAUCUGUUUGCCUUCAGAGUUGCGGCGUGCGGCUUGUGCGGGCUGCCCGCAUGCAAAGG ...-......((((((.(((((((((((....)))).(((.(((((((....))))))))))........))))))).))))(((((((...)))))))))..... ( -49.80) >DroEre_CAF1 37152 86 + 1 UGGUUUGGGGGCGCG--------------------GUGAGCGGCGAACAUCUGUUUGCCUUCAAAGUUGUGCCGUGCGGCUUGUGCGGGCUGUCCGCAUGCAAAGA ..((.((((((((((--------------------.((((.(((((((....)))))))))))....))))))..(((((((....))))))))).)).))..... ( -33.80) >DroYak_CAF1 40238 104 + 1 UGGCUUGGAGGCGGGGGGGACAC--CCAUUUGCGAGUGAGCGGCGAACAUCUGUUUGCCUUCAAAGUUGCGCCGUGCGGCUUGUGCGGGCUGCCCGCAUGCAAAGG ..(((((.(((.((((....).)--)).))).)))))(((.(((((((....))))))))))....(((((((....)))..(((((((...)))))))))))... ( -45.70) >DroPer_CAF1 47353 97 + 1 AGG-A---CGGCGGGUUGGGAGC-----AUUGCAAGUGAGAGGCAAACAUCUGUUUGCCUUCAAAGUUGCCUCGUGUGGCUUAUGCGGGCUACCAGCAUCCCACAC ...-.---.((.(((((((.(((-----.(((((..(((.((((((((....)))))))))))((((..(.....)..)))).)))))))).))))).)))).... ( -37.50) >consensus UGG_UUAGGGGCGGGUGGCAUGC__CCGUGUGCGAGUGAGCGGCGAACAUCUGUUUGCCUUCAAAGUUGCGGCGUGCGGCUUGUGCGGGCUGCCCGCAUGCAAAGG ..........((((((((..................((((.(((((((....)))))))))))(((((((.....))))))).......))))))))......... (-27.54 = -28.18 + 0.64)

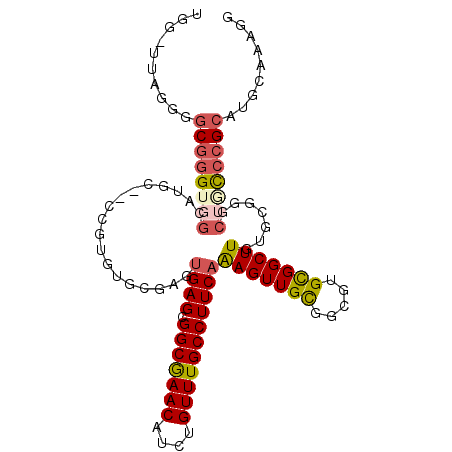
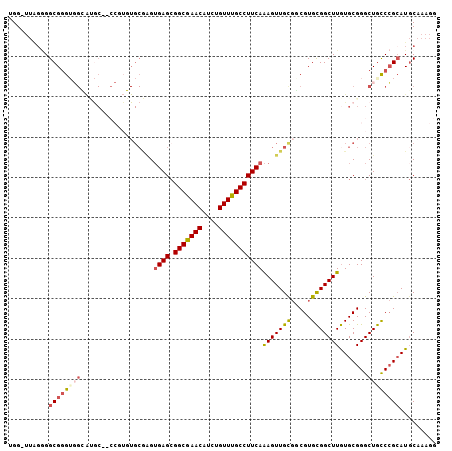
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:18 2006