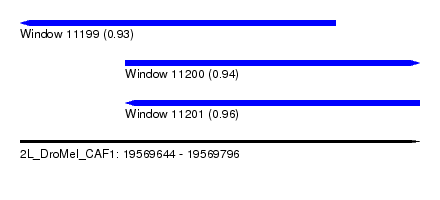
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,569,644 – 19,569,796 |
| Length | 152 |
| Max. P | 0.955535 |

| Location | 19,569,644 – 19,569,764 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19569644 120 - 22407834 UCCUCGAAAAACAUCGAUGUUUGUGAUUUUUUGAAAAAAUCGACGACAUCGACGUUGGCAUUUCAGUAAAGGAAUUCCAAAAUCUCAUUUACCUGUGAAAACCGCGCCCCAAAUGUUUUA .......((((((((((((((..(((((((.....)))))))..))))))))....(((......((((((..(((....)))..).)))))..(((.....)))))).....)))))). ( -26.00) >DroSec_CAF1 8965 108 - 1 UCCUCAAAAAACAUCGAUGUUUGUGAUUUUCCGAAAACAUCGAUGACAUCGAUGUUGCGAUUGCAACGAAGGAAUUCCAAAAUCUGAUUUGCCUGUAACAACGGCGCG------------ (((((......(((((((((((.((......)).)))))))))))........(((((....)))))).)))).................((((((....)))).)).------------ ( -30.10) >DroSim_CAF1 9556 120 - 1 UCCUCAAAAAACAUCGAUGUUUGUGAUUUUGCGAAAACAUCGAUGACAUCGAUGUUGCUAUUGCAACGAAGGAAUUCCAAAAUAUGAUUUGCCUGUGACAACGGCGCCCCAAAUGUUAUA (((((......(((((((((((.((......)).)))))))))))........(((((....)))))).)))).......(((((.....((((((....)))).)).....)))))... ( -32.80) >consensus UCCUCAAAAAACAUCGAUGUUUGUGAUUUUCCGAAAACAUCGAUGACAUCGAUGUUGCCAUUGCAACGAAGGAAUUCCAAAAUCUGAUUUGCCUGUGACAACGGCGCCCCAAAUGUU_UA ((((.....((((((((((((..((((.((.....)).))))..))))))))))))((....)).....)))).................((((((....)))).))............. (-22.04 = -22.60 + 0.56)


| Location | 19,569,684 – 19,569,796 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.97 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19569684 112 + 22407834 UUGGAAUUCCUUUACUGAAAUGCCAACGUCGAUGUCGUCGAUUUUUUCAAAAAAUCACAAACAUCGAUGUUUUUCGAGGAAUCAUCGAUGUUUCUCCACCUCUAAUAAGGC---G ..((....))...........((((((((((((((.((.((((((.....))))))))..)))))))))))....(((((((.......)))))))............)))---. ( -28.20) >DroSec_CAF1 8993 115 + 1 UUGGAAUUCCUUCGUUGCAAUCGCAACAUCGAUGUCAUCGAUGUUUUCGGAAAAUCACAAACAUCGAUGUUUUUUGAGGAAUCAUCGAUCUUUCUCCGCCUCUAAUGUGGCCUCU ..((((((((...(((((....))))).((((...(((((((((((...(.....)..)))))))))))....)))))))))....((....)))))(((.(....).))).... ( -33.90) >DroSim_CAF1 9596 112 + 1 UUGGAAUUCCUUCGUUGCAAUAGCAACAUCGAUGUCAUCGAUGUUUUCGCAAAAUCACAAACAUCGAUGUUUUUUGAGGAAUCAUCGAUCUUUCUCCGCCUCUAAUGUGGC---G ..((((((((...(((((....))))).((((...(((((((((((............)))))))))))....)))))))))....((....)))))(((.(....).)))---. ( -33.60) >consensus UUGGAAUUCCUUCGUUGCAAUAGCAACAUCGAUGUCAUCGAUGUUUUCGAAAAAUCACAAACAUCGAUGUUUUUUGAGGAAUCAUCGAUCUUUCUCCGCCUCUAAUGUGGC___G ..((((((((...(((((....))))).((((...(((((((((((............)))))))))))....)))))))))....((....)))))(((........))).... (-24.74 = -25.97 + 1.23)


| Location | 19,569,684 – 19,569,796 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.53 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19569684 112 - 22407834 C---GCCUUAUUAGAGGUGGAGAAACAUCGAUGAUUCCUCGAAAAACAUCGAUGUUUGUGAUUUUUUGAAAAAAUCGACGACAUCGACGUUGGCAUUUCAGUAAAGGAAUUCCAA (---(((((....))))))........((((.......))))..(((.((((((((..(((((((.....)))))))..)))))))).)))((.(((((......))))).)).. ( -30.00) >DroSec_CAF1 8993 115 - 1 AGAGGCCACAUUAGAGGCGGAGAAAGAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUCCGAAAACAUCGAUGACAUCGAUGUUGCGAUUGCAACGAAGGAAUUCCAA ....(((........)))((((....(((((((.((......))..(((((((((((.((......)).))))))))))).)))))))(((((....))))).......)))).. ( -34.80) >DroSim_CAF1 9596 112 - 1 C---GCCACAUUAGAGGCGGAGAAAGAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUGCGAAAACAUCGAUGACAUCGAUGUUGCUAUUGCAACGAAGGAAUUCCAA .---(((........)))((((....(((((((.((......))..(((((((((((.((......)).))))))))))).)))))))(((((....))))).......)))).. ( -35.80) >consensus C___GCCACAUUAGAGGCGGAGAAAGAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUCCGAAAACAUCGAUGACAUCGAUGUUGCCAUUGCAACGAAGGAAUUCCAA ....(((........)))((((....(((((((.((.....))...(((((((((((.((......)).))))))))))).)))))))(((((....))))).......)))).. (-27.86 = -27.53 + -0.32)

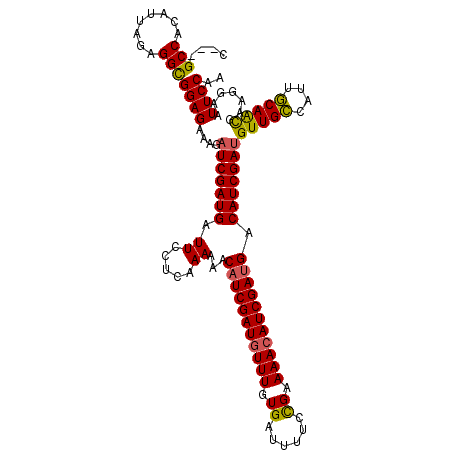
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:27 2006