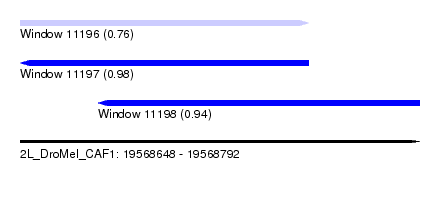
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,568,648 – 19,568,792 |
| Length | 144 |
| Max. P | 0.984504 |

| Location | 19,568,648 – 19,568,752 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -21.98 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
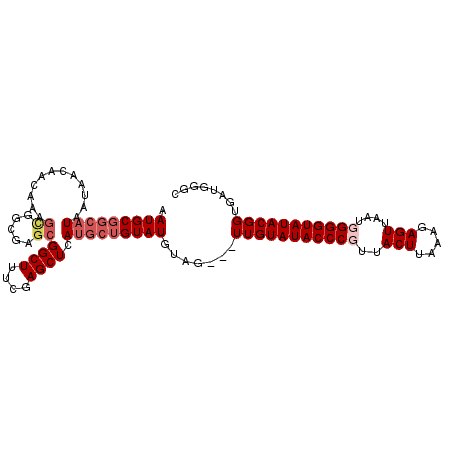
>2L_DroMel_CAF1 19568648 104 + 22407834 AAUGCGGCAUAAUAACAACAAAGUGGCGAGCGGCUUUCGAGCUCAUGCUGUAUG---UUGUUGUAUACCCGUUACUUGGAGAGUUAAUUGGGUAUACGGUGAUGGGC .(((((((((....((......))...((((.........)))))))))))))(---(..(((((((((((..((((...))))....)))))))))))..)).... ( -34.30) >DroSec_CAF1 8007 100 + 1 AAUGCGGCAUAAUAACAACAAU-------GCGGCUUUCGAGCUCAUGCUGUAUGUAGUUGUUGUAUACCCGUUACUGGGAGAGUUAAUUGGGUAUACGGUGAUGGGC ...((.((((..........))-------)).))......((((.(((.....)))((..(((((((((((..(((.....)))....)))))))))))..)))))) ( -30.20) >DroEre_CAF1 9709 103 + 1 -AUGCGGCAUAAUAACAACAAAGCGGCGAGCGGCUUUCGAGCUCAUGCUGUAUGUAG---UUGUAUACCCGUUACUUAAAUAGUUAAUGGGGUAUACGGUGAUGGGC -(((((((((............((.....))((((....)))).)))))))))((..---((((((((((((((((.....)).)))).))))))))))..)).... ( -29.30) >DroYak_CAF1 9025 94 + 1 AAUGCGGCAU---AACAACAAAGCGGCGAGCGGCUUUCGAGCUCAC---GUAUGUAG---UUGUAUACCCGUUACUUAAAUAGUUAUUGGGGUAUACGGUG----CC .....(((((---(((.(((..(((..((((.........)))).)---)).))).)---))((((((((...(((.....))).....)))))))).)))----)) ( -29.90) >consensus AAUGCGGCAUAAUAACAACAAAGCGGCGAGCGGCUUUCGAGCUCAUGCUGUAUGUAG___UUGUAUACCCGUUACUUAAAGAGUUAAUGGGGUAUACGGUGAUGGGC .(((((((((............((.....))((((....)))).))))))))).......(((((((((((..(((.....)))....)))))))))))........ (-21.98 = -23.85 + 1.87)

| Location | 19,568,648 – 19,568,752 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19568648 104 - 22407834 GCCCAUCACCGUAUACCCAAUUAACUCUCCAAGUAACGGGUAUACAACAA---CAUACAGCAUGAGCUCGAAAGCCGCUCGCCACUUUGUUGUUAUUAUGCCGCAUU ((........((((((((.....(((.....)))...))))))))(((((---((....((..((((.(....)..)))))).....)))))))........))... ( -27.10) >DroSec_CAF1 8007 100 - 1 GCCCAUCACCGUAUACCCAAUUAACUCUCCCAGUAACGGGUAUACAACAACUACAUACAGCAUGAGCUCGAAAGCCGC-------AUUGUUGUUAUUAUGCCGCAUU ((........((((((((.....(((.....)))...))))))))...........((((((...((.(....)..))-------..)))))).........))... ( -20.90) >DroEre_CAF1 9709 103 - 1 GCCCAUCACCGUAUACCCCAUUAACUAUUUAAGUAACGGGUAUACAA---CUACAUACAGCAUGAGCUCGAAAGCCGCUCGCCGCUUUGUUGUUAUUAUGCCGCAU- ((........((((((((.....(((.....)))...))))))))((---(.(((...(((.(((((.(....)..)))))..))).))).)))........))..- ( -26.60) >DroYak_CAF1 9025 94 - 1 GG----CACCGUAUACCCCAAUAACUAUUUAAGUAACGGGUAUACAA---CUACAUAC---GUGAGCUCGAAAGCCGCUCGCCGCUUUGUUGUU---AUGCCGCAUU ((----((..((((((((.....(((.....)))...))))))))((---(.(((...---((((((.(....)..)))))).....))).)))---.))))..... ( -30.80) >consensus GCCCAUCACCGUAUACCCAAUUAACUAUCCAAGUAACGGGUAUACAA___CUACAUACAGCAUGAGCUCGAAAGCCGCUCGCCGCUUUGUUGUUAUUAUGCCGCAUU ..........((((((((.....(((.....)))...))))))))..................((((.(....)..)))).......(((.((......)).))).. (-18.25 = -18.75 + 0.50)


| Location | 19,568,676 – 19,568,792 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -20.99 |
| Energy contribution | -23.30 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19568676 116 - 22407834 AUUAUGCAAAGUGUAUUUUGGAAAAAUUCUUUGCCUAAGAGCCCAUCACCGUAUACCCAAUUAACUCUCCAAGUAACGGGUAUACAACAA---CAUACAGCAUGAGCUCGAAAGCCGCU .((((((....(((((.(((((....(((((.....)))))....))...((((((((.....(((.....)))...))))))))..)))---.)))))))))))((.(....)..)). ( -26.00) >DroSec_CAF1 8029 118 - 1 GUUAUGCAAAGUGUAUGUUUGAAAAAUACUUUGUCUAAGGGCCCAUCACCGUAUACCCAAUUAACUCUCCCAGUAACGGGUAUACAACAACUACAUACAGCAUGAGCUCGAAAGCCGC- .((((((....((((((((((.................((........))((((((((.....(((.....)))...))))))))..)))..)))))))))))))(((....)))...- ( -27.10) >DroEre_CAF1 9736 116 - 1 CUUAUGCUAAGUAUUUGUUUGAAAAAUACUUUGUGUGAGGGCCCAUCACCGUAUACCCCAUUAACUAUUUAAGUAACGGGUAUACAA---CUACAUACAGCAUGAGCUCGAAAGCCGCU ((((((((((((((((.......)))))))).(((((.((........))((((((((.....(((.....)))...))))))))..---...)))))))))))))..(....)..... ( -33.50) >DroYak_CAF1 9050 105 - 1 CUUAUGCAAAGUAUUUGAUUAAAAAAAUC----UGUGAGGGG----CACCGUAUACCCCAAUAACUAUUUAAGUAACGGGUAUACAA---CUACAUAC---GUGAGCUCGAAAGCCGCU ..........((((..((((.....))))----.(((.((..----..))((((((((.....(((.....)))...))))))))..---.)))))))---(((.(((....)))))). ( -21.90) >consensus CUUAUGCAAAGUAUAUGUUUGAAAAAUACUUUGUCUAAGGGCCCAUCACCGUAUACCCAAUUAACUAUCCAAGUAACGGGUAUACAA___CUACAUACAGCAUGAGCUCGAAAGCCGCU .(((((((((((((((.......)))))))))))))))((((........((((((((.....(((.....)))...))))))))........(((.....))).)))).......... (-20.99 = -23.30 + 2.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:25 2006