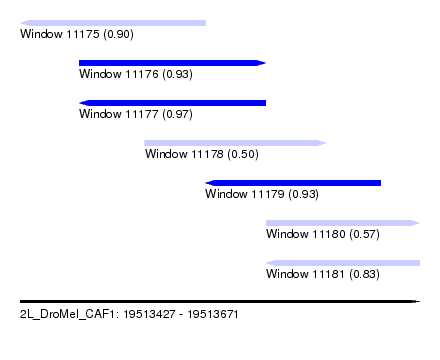
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,513,427 – 19,513,671 |
| Length | 244 |
| Max. P | 0.971336 |

| Location | 19,513,427 – 19,513,540 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -28.13 |
| Energy contribution | -27.80 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513427 113 - 22407834 ---ACACACUUACAUACUUGCUACACUGGUAUGUGUGCAAGUAUAUGUGCAAAAGCUUCGCCGCACAGUGGAACUGUCCAAUGGUUCC-CAAAAAACCAAUGUCCUUGUCCAUAAGA ---..........((((((((.((((......)))))))))))).(((((............)))))(((((...(..((.(((((..-.....))))).))..)...))))).... ( -33.20) >DroSec_CAF1 8676 117 - 1 CGCACACACUUACAUACUUGCUGCACUGGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCCGUCGCACAGUGGAACUCUCCAAUUGUUCUUUAAAAAAGCAAUGUCCUUGUCUAUAUGA .((...((((.((((((((((.((((......))))))))))))))((((............))))))))((((.........)))).........))................... ( -31.20) >DroSim_CAF1 8170 117 - 1 CGCACACACUUACAUACUCGCUGCACUAGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCGGUCGCACAGUGGAACUCUCCAAUUGUUCUUUAAAAAAGCAAUGUCCUUGUCCAUAUGA .(((((((......(((((((.((((......))))))))))))))))))....((......))...(((((........((((((.........)))))).......))))).... ( -34.86) >consensus CGCACACACUUACAUACUUGCUGCACUGGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCCGUCGCACAGUGGAACUCUCCAAUUGUUCUUUAAAAAAGCAAUGUCCUUGUCCAUAUGA ......((((.((((((((((.((((......))))))))))))))((((............))))))))((((.........)))).............................. (-28.13 = -27.80 + -0.33)

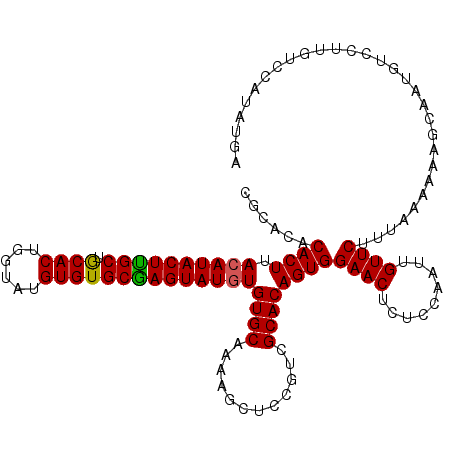
| Location | 19,513,463 – 19,513,577 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513463 114 + 22407834 UGGACAGUUCCACUGUGCGGCGAAGCUUUUGCACAUAUACUUGCACACAUACCAGUGUAGCAAGUAUGUAAGUGUGU------GAGUGCUUUCUCGCAGAGAGAGAAAAUAAAGAAAACG ((((....))))((.(((((.(((((.((..(((((((((((((((((......)))).))))))))....))))).------.)).))))).))))).))................... ( -42.50) >DroSec_CAF1 8713 118 + 1 UGGAGAGUUCCACUGUGCGACGGAGCUUUUGCACACAUACUCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGUGAGUGCUCUCUUGCAGA--GAGAAAAUAAAGAAAACG ..(((((((((..........)))))))))((((.(((((.(((((((.(((..((((.....))))))).))))))).))))).))))((((((...))--)))).............. ( -41.80) >DroSim_CAF1 8207 118 + 1 UGGAGAGUUCCACUGUGCGACCGAGCUUUUGCACACAUACUCGCACACAUACUAGUGCAGCGAGUAUGUAAGUGUGUGCGUGUAAGUGCUCUCUUACAGA--GAGAAAAUAAAGAAAUCG (((......)))(((((.((..((((.(((((((((((((((((.(((......)))..))))))))))..((....))))))))).)))))).))))).--.................. ( -40.10) >consensus UGGAGAGUUCCACUGUGCGACGGAGCUUUUGCACACAUACUCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGUGAGUGCUCUCUUGCAGA__GAGAAAAUAAAGAAAACG ((((....))))(((((.((..((((.(((((((((((((((((.(((......)))..))))))))))..........))))))).)))))).)))))..................... (-30.33 = -30.23 + -0.10)


| Location | 19,513,463 – 19,513,577 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.49 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513463 114 - 22407834 CGUUUUCUUUAUUUUCUCUCUCUGCGAGAAAGCACUC------ACACACUUACAUACUUGCUACACUGGUAUGUGUGCAAGUAUAUGUGCAAAAGCUUCGCCGCACAGUGGAACUGUCCA .((((......(((((((.......))))))).....------.(((......((((((((.((((......)))))))))))).(((((............))))))))))))...... ( -30.00) >DroSec_CAF1 8713 118 - 1 CGUUUUCUUUAUUUUCUC--UCUGCAAGAGAGCACUCACACGCACACACUUACAUACUUGCUGCACUGGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCCGUCGCACAGUGGAACUCUCCA .((((..........(((--((.....)))))......((((((((((......(((((((.((((......))))))))))))))))))....((......))...)))))))...... ( -35.00) >DroSim_CAF1 8207 118 - 1 CGAUUUCUUUAUUUUCUC--UCUGUAAGAGAGCACUUACACGCACACACUUACAUACUCGCUGCACUAGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCGGUCGCACAGUGGAACUCUCCA .((.(((........(((--((.....)))))......((((((((((......(((((((.((((......))))))))))))))))))....((......))...)))))).)).... ( -37.90) >consensus CGUUUUCUUUAUUUUCUC__UCUGCAAGAGAGCACUCACACGCACACACUUACAUACUUGCUGCACUGGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCCGUCGCACAGUGGAACUCUCCA .((((((((................)))))))).............((((.((((((((((.((((......))))))))))))))((((............)))))))).......... (-30.93 = -30.49 + -0.44)

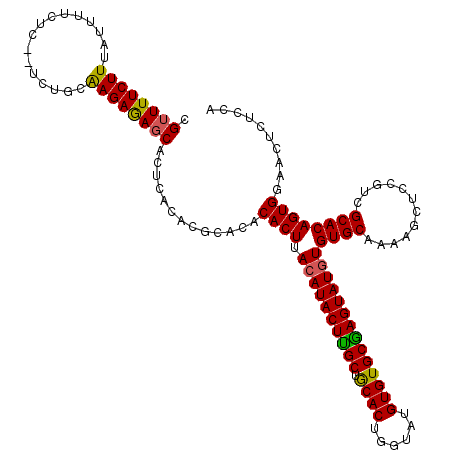
| Location | 19,513,503 – 19,513,614 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513503 111 + 22407834 UUGCACACAUACCAGUGUAGCAAGUAUGUAAGUGUGU------GAGUGCUUUCUCGCAGAGAGAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGCAAAACCAACACACAUAUA---U ((((((((......)))).))))..(((((.((((((------...(((((((((.....)))))).......((.((((((....)).)))).)))))......)))))).))))---) ( -30.20) >DroSec_CAF1 8753 114 + 1 UCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGUGAGUGCUCUCUUGCAGA--GAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGUAAAAUCAACA----UAUAAAUA ..((((((((....((((.....))))....))))))))((((((.(((((((((...))--)))).......((.((((((....)).)))).)))))...)).)))----)....... ( -28.90) >DroSim_CAF1 8247 118 + 1 UCGCACACAUACUAGUGCAGCGAGUAUGUAAGUGUGUGCGUGUAAGUGCUCUCUUACAGA--GAGAAAAUAAAGAAAUCGUAUCGUUAUCGUUAUCGUAAAAUCAAAAUAAAUAUAGAUA .(((((((((....((((.....))))....)))))))))((((....(((((.....))--)))........((...((((.((....)).)).)).....))........)))).... ( -23.80) >consensus UCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGUGAGUGCUCUCUUGCAGA__GAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGUAAAAUCAACA_A_AUAUA_AUA .(((((((((....((((.....))))....)))))))))......(((..((((.......)))).......((.((((((....)).)))).)))))..................... (-16.76 = -17.43 + 0.67)

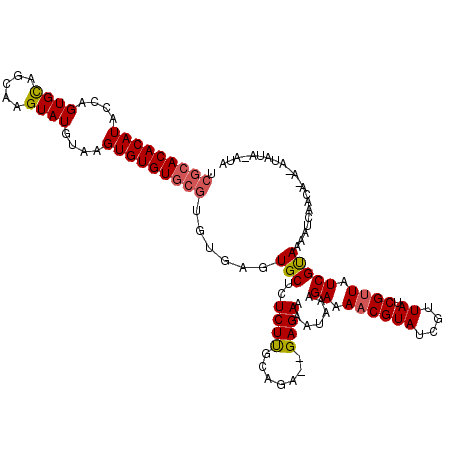
| Location | 19,513,540 – 19,513,647 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.56 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513540 107 - 22407834 ACAAUCGUUAUCAAAAAACCAGAUAUGCUUGUAA---UAUAUGUGUGUUGGUUUUGCGAUAACGAUAACGAUACGUUUUCUUUAUUUUCUCUCUCUGCGAGAAAGCACUC--- ...(((((((((..(((((((.((((((......---.....)))))))))))))..)))))))))........((((((((................))))))))....--- ( -27.59) >DroSec_CAF1 8793 106 - 1 GCAAUCGUUAUCAAAAAG-CAGAUAUGGAUAUAUAUUUAUA----UGUUGAUUUUACGAUAACGAUAACGAUACGUUUUCUUUAUUUUCUC--UCUGCAAGAGAGCACUCACA ((.(((((((((..((((-(((((((((((....)))))))----).))).))))..))))))))).....................((((--(.....)))))))....... ( -22.00) >DroSim_CAF1 8287 110 - 1 GCUAUCGUUAUCAAAAAA-CAGAUAUGCAUAUAUAUCUAUAUUUAUUUUGAUUUUACGAUAACGAUAACGAUACGAUUUCUUUAUUUUCUC--UCUGUAAGAGAGCACUUACA ((((((((((((......-.(((((((....)))))))....(((((.((....)).))))).))))))))))..............((((--(.....)))))))....... ( -21.40) >consensus GCAAUCGUUAUCAAAAAA_CAGAUAUGCAUAUAUAU_UAUAU_U_UGUUGAUUUUACGAUAACGAUAACGAUACGUUUUCUUUAUUUUCUC__UCUGCAAGAGAGCACUCACA ...(((((((((..((((...((((((..((((....))))..))))))..))))..)))))))))........((((((((................))))))))....... (-17.21 = -17.56 + 0.34)

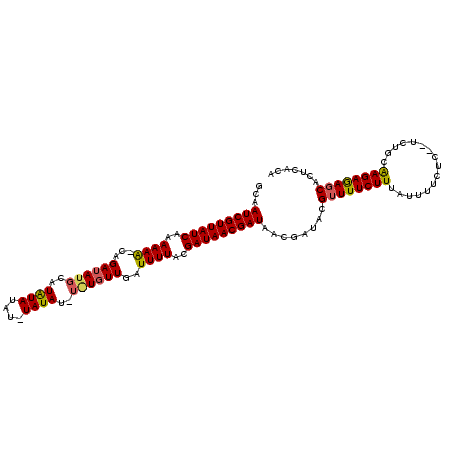
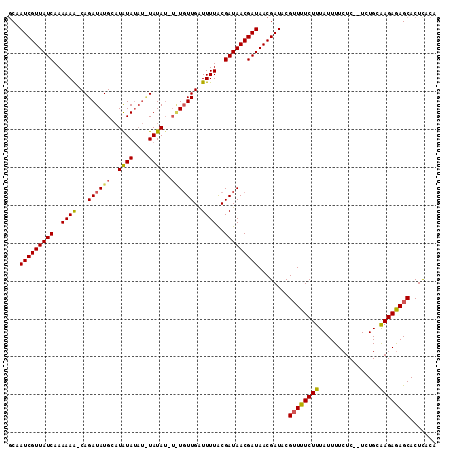
| Location | 19,513,577 – 19,513,671 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -17.59 |
| Consensus MFE | -10.32 |
| Energy contribution | -10.76 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513577 94 + 22407834 UAUCGUUAUCGUUAUCGCAAAACCAACACACAUAUA---UUACAAGCAUAUCUGGUUUUUUGAUAACGAUUGUCACAUAAAGUUAUUGCUCCUUAAC ....(..((((((((((.(((((((...........---.............))))))).))))))))))...).......((((........)))) ( -17.36) >DroSec_CAF1 8831 92 + 1 UAUCGUUAUCGUUAUCGUAAAAUCAACA----UAUAAAUAUAUAUCCAUAUCUG-CUUUUUGAUAACGAUUGCCACAUAAAGUUUUUGCUCUUUAAC ....((.((((((((((.((((.((..(----(((..((....))..)))).))-.)))))))))))))).))....(((((........))))).. ( -13.50) >DroSim_CAF1 8325 96 + 1 UAUCGUUAUCGUUAUCGUAAAAUCAAAAUAAAUAUAGAUAUAUAUGCAUAUCUG-UUUUUUGAUAACGAUAGCCACAUAAAGUUAUUGCUCUUUAAC ....(((((((((((((.((((...........((((((((......)))))))-))))))))))))))))))....(((((........))))).. ( -21.90) >consensus UAUCGUUAUCGUUAUCGUAAAAUCAACA_A_AUAUA_AUAUAUAUGCAUAUCUG_UUUUUUGAUAACGAUUGCCACAUAAAGUUAUUGCUCUUUAAC ....((.((((((((((.((((.((...........................)).)))).)))))))))).))....(((((........))))).. (-10.32 = -10.76 + 0.45)

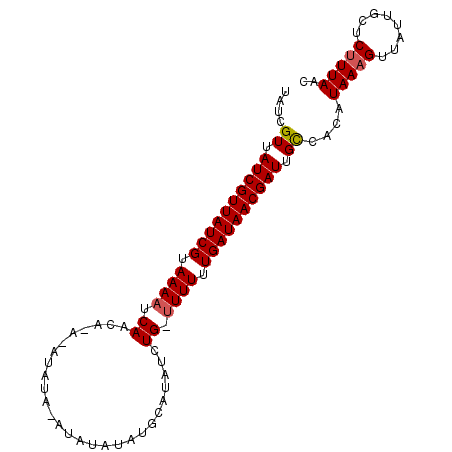
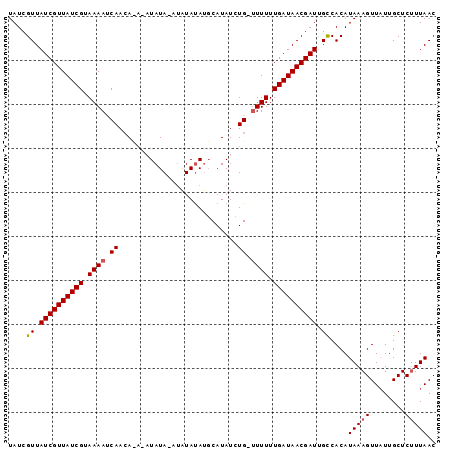
| Location | 19,513,577 – 19,513,671 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.17 |
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19513577 94 - 22407834 GUUAAGGAGCAAUAACUUUAUGUGACAAUCGUUAUCAAAAAACCAGAUAUGCUUGUAA---UAUAUGUGUGUUGGUUUUGCGAUAACGAUAACGAUA ((((.((((......))))...)))).(((((((((..(((((((.((((((......---.....)))))))))))))..)))))))))....... ( -24.60) >DroSec_CAF1 8831 92 - 1 GUUAAAGAGCAAAAACUUUAUGUGGCAAUCGUUAUCAAAAAG-CAGAUAUGGAUAUAUAUUUAUA----UGUUGAUUUUACGAUAACGAUAACGAUA ((((....((....((.....)).))..((((((((..((((-(((((((((((....)))))))----).))).))))..)))))))))))).... ( -18.50) >DroSim_CAF1 8325 96 - 1 GUUAAAGAGCAAUAACUUUAUGUGGCUAUCGUUAUCAAAAAA-CAGAUAUGCAUAUAUAUCUAUAUUUAUUUUGAUUUUACGAUAACGAUAACGAUA ..(((((........)))))(((.(.((((((((((..((((-((((.(((.(((((....))))).))))))).))))..)))))))))).).))) ( -17.90) >consensus GUUAAAGAGCAAUAACUUUAUGUGGCAAUCGUUAUCAAAAAA_CAGAUAUGCAUAUAUAU_UAUAU_U_UGUUGAUUUUACGAUAACGAUAACGAUA ((((.((((......))))...)))).(((((((((..((((...((((((..((((....))))..))))))..))))..)))))))))....... (-15.12 = -15.13 + 0.01)

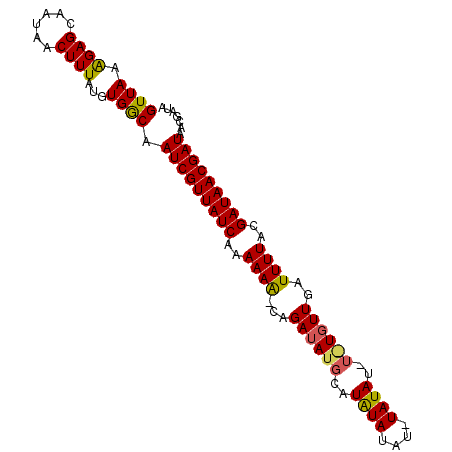
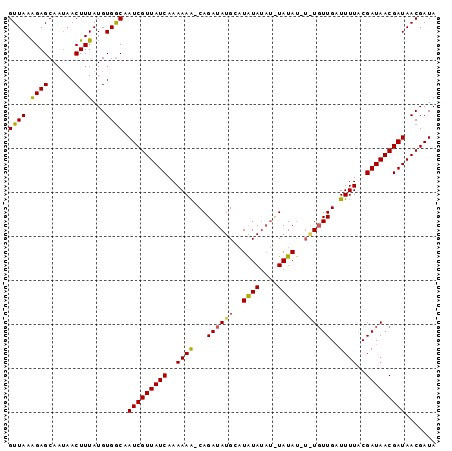
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:10 2006