| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,500,451 – 19,500,553 |
| Length | 102 |
| Max. P | 0.925821 |

| Location | 19,500,451 – 19,500,553 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.06 |
| Mean single sequence MFE | -32.27 |
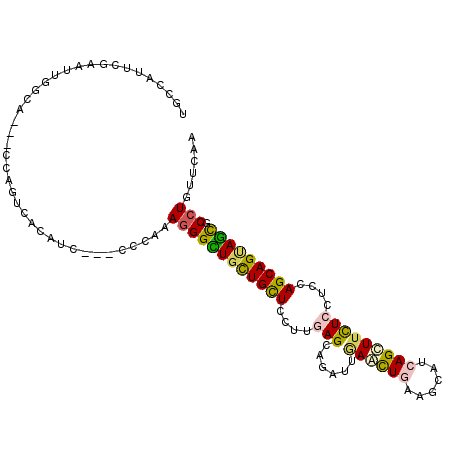
| Consensus MFE | -17.66 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19500451 102 + 22407834 UGCAACUGGAAUUGGUG---CCGGCCACAUC---UCCAAAGGGCUGCUGCUCCUUGAGCAAAUUGAAUUGAAGCAUCAGUUUCUCCUCCAGCAGUAGCGCCUGUUCAA ((.(((((((..((((.---...))))....---)))).((((((((((((....(((.(((((((.........))))))))))....))))))))).)))))))). ( -33.50) >DroVir_CAF1 18126 102 + 1 UUCCAUUCAUAUUGGCC---AUAUUGAGAUC---CCCAAACGGUUGUUGUUGCUUCAGCAGAUUGAACUGAAGCAACAGCUUUUCUUCCAGCAGCAGCGCUUGUUCGA ......((..((.(((.---.....((((..---.((....))..(((((((((((((.........)))))))))))))...))))...((....))))).))..)) ( -24.20) >DroGri_CAF1 17697 105 + 1 UGCCAUUCGAAUUGGCAUUGGUAUUGAGAUC---CCCGAAGGGCUGCUGCUGCUUGAGCAGAUUGAAUUGCAGCAGCAGCUUCUCCUCCAGCAGAAGCGCCUGUUCGA ......((((((.(((..(((....((((..---((....))(((((((((((.....(.....)....))))))))))).))))..)))((....))))).)))))) ( -42.40) >DroYak_CAF1 15304 102 + 1 UGCAAUUGGAAUUGCUG---CCAGCCACAUC---UCCAAAGGGCUGCUGCUCCUUAAGCAAAUUAAACUGAAGCAUAAGUUUCUCCUCCAGCAGUAGCGCCUGUUCAA .....(((((..((.((---.....))))..---)))))((((((((((((..((((.....))))...(((((....)))))......))))))))).)))...... ( -27.40) >DroMoj_CAF1 19068 105 + 1 UGCCAUUCGUGUUAGCC---GCAUUGAGAUCUUCGCCGAAGGGUUGUUGCUGCUUGAGCAGAUUGAACUGUAGCAUCAGUUUCUCCUCCAGCAGUAACGCCUGUUCAA .....(((((((.....---))).)))).........(((.((.((((((((((.(((.(((...(((((......)))))))).))).)))))))))).)).))).. ( -32.10) >DroAna_CAF1 15452 102 + 1 UACCGCCAUUAGCGGUA---CCAGUCACAUC---GCCAAAGGGUUGCUGCUCCUUGAGCAGAUUCAGCUGCAACAUAAGCUUCUCCUCCAGCAGCAAUGCCUGCUCGA ((((((.....))))))---.........((---(....((((((((((((....(((.(((...((((........))))))).))).))))))))).)))...))) ( -34.00) >consensus UGCCAUUCGAAUUGGCA___CCAGUCACAUC___CCCAAAGGGCUGCUGCUCCUUGAGCAGAUUGAACUGAAGCAUCAGCUUCUCCUCCAGCAGUAGCGCCUGUUCAA .......................................((((((((((((....(((......((((((......)))))))))....))))))))).)))...... (-17.66 = -17.38 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:01 2006