
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,500,182 – 19,500,291 |
| Length | 109 |
| Max. P | 0.943246 |

| Location | 19,500,182 – 19,500,291 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -44.93 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
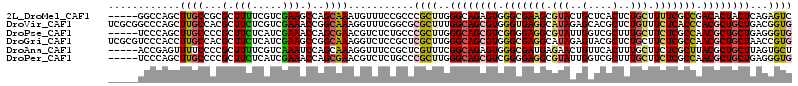
>2L_DroMel_CAF1 19500182 109 + 22407834 -----GGCCAGCUUGCCGCGCUUUUCGUCGAAGCCAGCAAAUGUUUCCGCCCGCUUGGGCAGAGUGGGCGAAGCGUACUGCUCACUCUGCUUUUCGCCGACACUACUCAGAGUC -----(((.(((((((.(.(((((.....)))))).))))........((((....))))((((((((((........)))))))))))))....)))(((.((....)).))) ( -37.70) >DroVir_CAF1 17852 114 + 1 UCGCGGCCCAGCUUGCCACGCUUCUCGUCGAAACCGGCAAAGGUUUCGGCGCGCUUUGGCAGCGUGGGUGAGGCAUAGAGCACGCUCUGUUUCUCACCCACGCUGCUGACGGUG .((.(((.......))).)).....((((((((((......))))))))))(((((..(((((((((((((((.((((((....)))))).)))))))))))))))..).)))) ( -65.90) >DroPse_CAF1 18447 109 + 1 -----UCCCAGCUUGCCCCGCUUCUCAUCGAAACCAGCGAACGUCUCUGCCCGCUUGGGCAGCGUCGGGGAGGCGUAUUGGUCGCUUUGCUUCUCGCCAACGCUGCUGAGGGUG -----(((((((..((..((((((((.(((.......))).((.(.((((((....)))))).).))))))))))..((((.((..........)))))).)).)))).))).. ( -41.20) >DroGri_CAF1 17423 114 + 1 UCGCGUCCCACCUUGCCACGCUUCUCAUCGAAGCCGGCAAAGGUCUCCGCUCGCUUGGGCAGCGUGGGCGAGGCAUAGAGUACGCUCUGCUUCUCGCCAACGCUGCUAACCGUG ..(((....((((((((..(((((.....))))).))).)))))...))).(((...((((((((.(((((((..(((((....)))))..))))))).))))))))....))) ( -55.20) >DroAna_CAF1 15183 109 + 1 -----ACCGAGUUUUCCCCGCUUUUCGUCAAAUCCAGCAAAGGUUUCCGCUCGUUUCGGCAGAGUGGGCGAUGAGAACUGUUCACUUUGCUUCUCGCUUACGCUGCUUAGUGCU -----..(((((.......((((((.((........))))))))....)))))....(((((.((((((((.(((....(....)....))).)))))))).)))))....... ( -28.40) >DroPer_CAF1 16505 109 + 1 -----UCCCAGCUUGCCCCGCUUCUCAUCGAAACCAGCGAACGUCUCUGCCCGCUUGGGCAGCGUCGGGGAGGCGUAUUGGUCGCUUUGCUUCUCGCCAACGCUGCUGAGGGUG -----(((((((..((..((((((((.(((.......))).((.(.((((((....)))))).).))))))))))..((((.((..........)))))).)).)))).))).. ( -41.20) >consensus _____UCCCAGCUUGCCCCGCUUCUCAUCGAAACCAGCAAAGGUCUCCGCCCGCUUGGGCAGCGUGGGCGAGGCGUACUGCUCGCUCUGCUUCUCGCCAACGCUGCUGAGGGUG ............((((...(.(((.....))).)..))))...........((((..((((((((.(((((((.(((..(....)..))).))))))).))))))))...)))) (-23.31 = -23.78 + 0.48)


| Location | 19,500,182 – 19,500,291 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -46.62 |
| Consensus MFE | -25.33 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
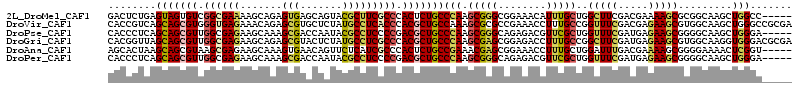
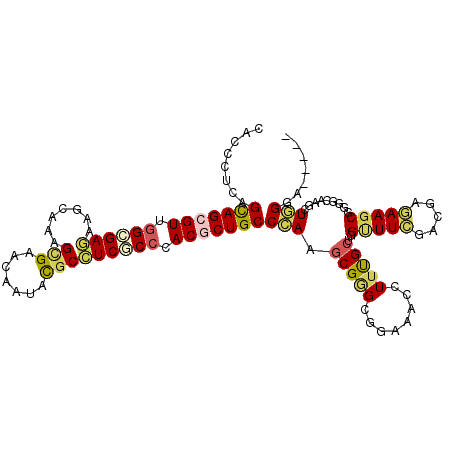
>2L_DroMel_CAF1 19500182 109 - 22407834 GACUCUGAGUAGUGUCGGCGAAAAGCAGAGUGAGCAGUACGCUUCGCCCACUCUGCCCAAGCGGGCGGAAACAUUUGCUGGCUUCGACGAAAAGCGCGGCAAGCUGGCC----- (((.((....)).)))........((((((((.((((....))..)).))))))))....((.((((....)..((((((((((.......)))).))))))))).)).----- ( -39.80) >DroVir_CAF1 17852 114 - 1 CACCGUCAGCAGCGUGGGUGAGAAACAGAGCGUGCUCUAUGCCUCACCCACGCUGCCAAAGCGCGCCGAAACCUUUGCCGGUUUCGACGAGAAGCGUGGCAAGCUGGGCCGCGA ...((((.((((((((((((((..(.((((....)))).)..))))))))))))))...........((((((......)))))))))).....((((((.......)))))). ( -58.30) >DroPse_CAF1 18447 109 - 1 CACCCUCAGCAGCGUUGGCGAGAAGCAAAGCGACCAAUACGCCUCCCCGACGCUGCCCAAGCGGGCAGAGACGUUCGCUGGUUUCGAUGAGAAGCGGGGCAAGCUGGGA----- ....((((((((((((((.(((.......(((.......)))))).))))))))((((.(((((((......))))))).(((((.....))))).))))..)))))).----- ( -49.41) >DroGri_CAF1 17423 114 - 1 CACGGUUAGCAGCGUUGGCGAGAAGCAGAGCGUACUCUAUGCCUCGCCCACGCUGCCCAAGCGAGCGGAGACCUUUGCCGGCUUCGAUGAGAAGCGUGGCAAGGUGGGACGCGA ..((.((.(((((((.((((((..(.((((....)))).)..)))))).)))))))..)).)).(((....((((((((((((((.....))))).)))))))).)...))).. ( -55.30) >DroAna_CAF1 15183 109 - 1 AGCACUAAGCAGCGUAAGCGAGAAGCAAAGUGAACAGUUCUCAUCGCCCACUCUGCCGAAACGAGCGGAAACCUUUGCUGGAUUUGACGAAAAGCGGGGAAAACUCGGU----- .((.....((((.((..((((..((.(..(....)..).))..))))..)).))))((((.(.((((((....)))))).).)))).......))(((.....)))...----- ( -27.50) >DroPer_CAF1 16505 109 - 1 CACCCUCAGCAGCGUUGGCGAGAAGCAAAGCGACCAAUACGCCUCCCCGACGCUGCCCAAGCGGGCAGAGACGUUCGCUGGUUUCGAUGAGAAGCGGGGCAAGCUGGGA----- ....((((((((((((((.(((.......(((.......)))))).))))))))((((.(((((((......))))))).(((((.....))))).))))..)))))).----- ( -49.41) >consensus CACCCUCAGCAGCGUUGGCGAGAAGCAAAGCGAACAAUACGCCUCGCCCACGCUGCCCAAGCGGGCGGAAACCUUUGCUGGUUUCGACGAGAAGCGGGGCAAGCUGGGA_____ ........(((((((.((((((.......(((.......))))))))).)))))))(((.(((((........)))))..(((((.....))))).........)))....... (-25.33 = -25.48 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:25:00 2006