
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,499,679 – 19,499,829 |
| Length | 150 |
| Max. P | 0.869683 |

| Location | 19,499,679 – 19,499,790 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.11 |
| Mean single sequence MFE | -41.11 |
| Consensus MFE | -21.12 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
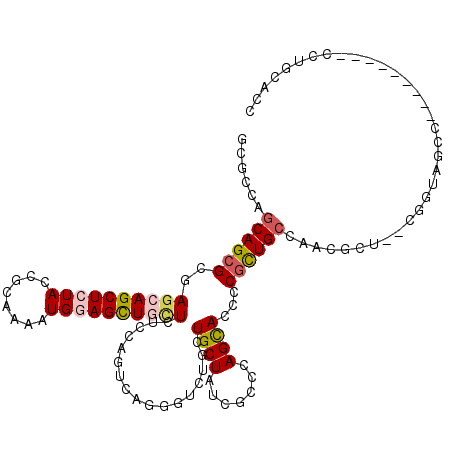
>2L_DroMel_CAF1 19499679 111 - 22407834 GCGCCAGCAGCGUGAACAGCUGUACAAGAAGAUGGAGCUCCUCUCCAGUCAGGGUCUGUUGCUGUCGCCCAGCACUCCCCUGCCACCG------GUCAGCAACAAUCAUCCUCCACC ......((((((....).)))))....((...(((((.....))))).))(((((.(((((((((((....(((......)))...))------).))))))))...)))))..... ( -33.40) >DroVir_CAF1 17255 108 - 1 GCGUCAACAGCGCGAGCAGCUAUAUCGCAAAAUGGAGCUCCUGUCCAGUCAGGGUCUGUUGCUAUCGCCCAGCACGCCGUUGCCAACGCUAUCGCCAGCG---------CUGGCGCC ((((...((((((..((.((......))....((((((..(((......)))(((.(((((........))))).)))))).)))........))..)))---------))))))). ( -35.40) >DroPse_CAF1 17932 108 - 1 GCGCCAGCAGCGGGAGCAGCUCUACCGCAAGAUGGAGCUGCUCUCUAGCCAGGGUCUUCUGCUCUCUCCCAGCACCCCGCUGCCGCCCAUGCCGGUGGCC---------UCUACUCC ..(((.((((((((((((((((((.(....).)))))))))))....((..(((.............))).))...))))))).(((......)))))).---------........ ( -48.72) >DroGri_CAF1 16844 96 - 1 GCGUCAGCAGCGCGAGCAGCUCUAUCGCAAAAUGGAGCUGCUGUCCAGUCAGGGAUUGCUGCUGUCGCCGAGCACACCGCUGCCGACAC---------------------UGGCACC (((.(((((((...(((((((((((......)))))))))))((((......)))).))))))).)))..(((.....)))((((....---------------------))))... ( -46.40) >DroMoj_CAF1 18188 108 - 1 ACGCCAGCAGCGUGAGCAGCUCUAUCGGAAAAUGGAGUUGUUGUCCAGUCAAGGCCUUUUGCUAUCGCCCAGUACUCCGCUGCCAACGCUUGCGCCAGCC---------CUCGCACC ..((..((.(((..(((((((((((......))))))))((((..((((..((..((...((....))..))..))..)))).)))))))..)))..)).---------...))... ( -34.00) >DroPer_CAF1 15990 108 - 1 GCGCCAGCAGCGGGAGCAGCUCUACCGCAAGAUGGAGCUGCUCUCUAGCCAGGGUCUUCUGCUCUCUCCCAGCACCCCGCUGCCGCCCAUGCCGGUGGCC---------UCUACUCC ..(((.((((((((((((((((((.(....).)))))))))))....((..(((.............))).))...))))))).(((......)))))).---------........ ( -48.72) >consensus GCGCCAGCAGCGCGAGCAGCUCUACCGCAAAAUGGAGCUGCUCUCCAGUCAGGGUCUGCUGCUAUCGCCCAGCACCCCGCUGCCAACGCU__CGGUAGCC_________CCUGCACC ......((((((..((((((((((........)))))))))).................((((.......))))...)))))).................................. (-21.12 = -21.77 + 0.64)


| Location | 19,499,712 – 19,499,829 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.54 |
| Mean single sequence MFE | -44.44 |
| Consensus MFE | -28.84 |
| Energy contribution | -28.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
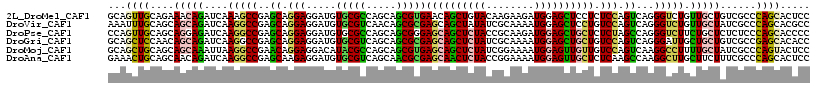
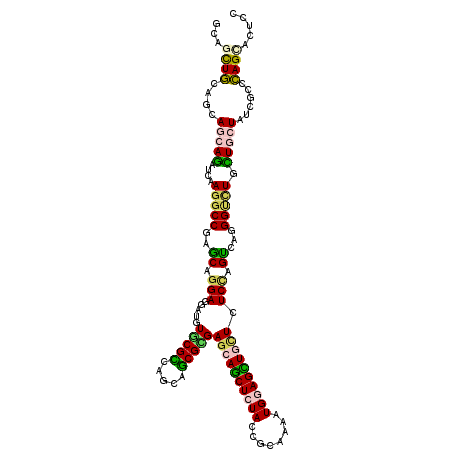
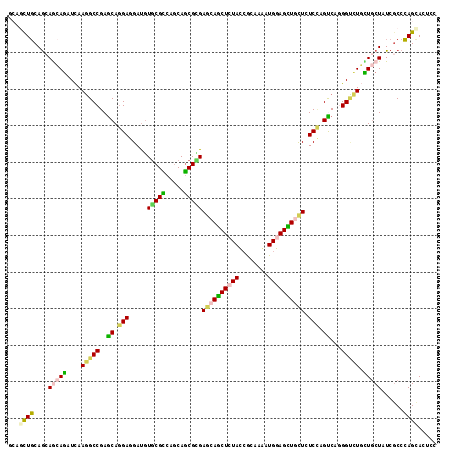
>2L_DroMel_CAF1 19499712 117 - 22407834 GCAGUUGCAGAAACAGAUCAAAGCCGAGCAGGAGGAUGUGCGCCAGCAGCGUGAACAGCUGUACAAGAAGAUGGAGCUCCUCUCCAGUCAGGGUCUGUUGCUGUCGCCCAGCACUCC ((.((.((((.((((((((...((.(.(..((((((...((.(((((((((....).))))).........))).)))))))))).))...)))))))).)))).))...))..... ( -36.30) >DroVir_CAF1 17285 117 - 1 AAAUUUGCAGCAGCAGAUCAAGGCCGAGCAGGAGGAUGUGCGUCAACAGCGCGAGCAGCUAUAUCGCAAAAUGGAGCUCCUGUCCAGUCAGGGUCUGUUGCUAUCGCCCAGCACGCC ......(((((((((((((..(((.(..((((((((((((((....).((....)).)).))))).(.....)...))))))..).)))..)))))))))))...((...))..)). ( -43.20) >DroPse_CAF1 17962 117 - 1 CCAGUUGCAGCAGGAGAUCAAGGCCGAGCAGGAGGAUGUGCGCCAGCAGCGGGAGCAGCUCUACCGCAAGAUGGAGCUGCUCUCUAGCCAGGGUCUUCUGCUCUCUCCCAGCACCCC ...((((.(((((((((((..(((...((.((.(......).)).))...(((((((((((((.(....).)))))))))))))..)))..)))))))))))......))))..... ( -54.70) >DroGri_CAF1 16862 117 - 1 GCAGCUCCAACAGCAGAUCAAGGCCGAGCAGGAGGAUGUGCGUCAGCAGCGCGAGCAGCUCUAUCGCAAAAUGGAGCUGCUGUCCAGUCAGGGAUUGCUGCUGUCGCCGAGCACACC ...((((.....(((.(((....((.....))..))).)))(.(((((((...(((((((((((......)))))))))))((((......)))).))))))).)...))))..... ( -51.80) >DroMoj_CAF1 18218 117 - 1 GCAGCUGCAGCAGCAAAUUAAGGCCGAACAGGAGGACAUACGCCAGCAGCGUGAGCAGCUCUAUCGGAAAAUGGAGUUGUUGUCCAGUCAAGGCCUUUUGCUAUCGCCCAGUACUCC ...((((..((((((((...(((((........((((.(((((.....)))))(((((((((((......)))))))))))))))......)))))))))))...)).))))..... ( -41.64) >DroAna_CAF1 14728 117 - 1 GAAACUGCAGCAACAGAUCAAGGCCGAGCAAGAGGAUGUGCGUCAGCAACGCGAGCAACUCUACCGGAAAAUGGAGUUGCUCUCAAGCCAAGGCUUGCUUCUUUCGCCCAGCACUCC ....(((......))).....((.(((..((((((....((((.....))))(((((((((((........))))))))))).(((((....)))))))))))))).))........ ( -39.00) >consensus GCAGCUGCAGCAGCAGAUCAAGGCCGAGCAGGAGGAUGUGCGCCAGCAGCGCGAGCAGCUCUACCGCAAAAUGGAGCUGCUCUCCAGUCAGGGUCUGCUGCUAUCGCCCAGCACUCC ...((((....(((((....(((((..((.(((.....(((((.....)))))((((((((((........)))))))))).))).))...))))).)))))......))))..... (-28.84 = -28.40 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:58 2006