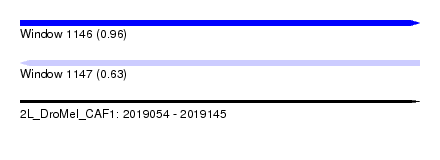
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,019,054 – 2,019,145 |
| Length | 91 |
| Max. P | 0.956047 |

| Location | 2,019,054 – 2,019,145 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -15.35 |
| Energy contribution | -17.41 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2019054 91 + 22407834 GAAGGGUGGGGGAAGUGGAAA-UGAAAAUGGAAGGGGAAAAGCCGCAUAUCCUGCUCAUGUAUGCAUGUGUGCGAGCCUAGGCAAGCAAAUA ...(((..(((...((((..(-(....)).............))))...)))..)))(((....)))...(((..((....))..))).... ( -20.96) >DroSec_CAF1 36580 84 + 1 GAAGGGUGGGGAAAGUGGAAA-UGAAA-------GGGAAAAGCCGCAUAUCCUGCUCAUGUAUGCAUGUGUGCGGCUGUAGGCAAGCAAAAU ...(((..(((...((((...-.....-------........))))...)))..))).....(((.(((.(((....))).))).))).... ( -22.49) >DroSim_CAF1 35024 84 + 1 AAAGGGUGGGGAAAGUGGAAA-UGGAA-------GGGAAAAGCCGCAUAUCCUGCUCAUGUAUGCAUGUGUGCGGCUGUAGGCAAGCAAAUA ...(((..(((...((((...-.....-------........))))...)))..))).....(((.(((.(((....))).))).))).... ( -22.49) >DroYak_CAF1 38230 73 + 1 ------------AAGUGGAAAGUGGAA-------GGAGAAAGCCGCAUAUCCUGCUCAUGUAUGCAUGUGUGCGAGCGUGGGCAAGCAAAUA ------------..(((((..((((..-------........))))...)))(((((((((.((((....)))).))))))))).))..... ( -23.30) >consensus GAAGGGUGGGGAAAGUGGAAA_UGAAA_______GGGAAAAGCCGCAUAUCCUGCUCAUGUAUGCAUGUGUGCGACCGUAGGCAAGCAAAUA ...((((((((...((((........................))))...)))))))).....(((.(((.(((....))).))).))).... (-15.35 = -17.41 + 2.06)

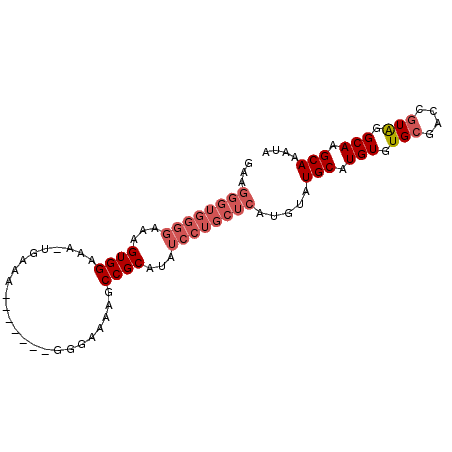
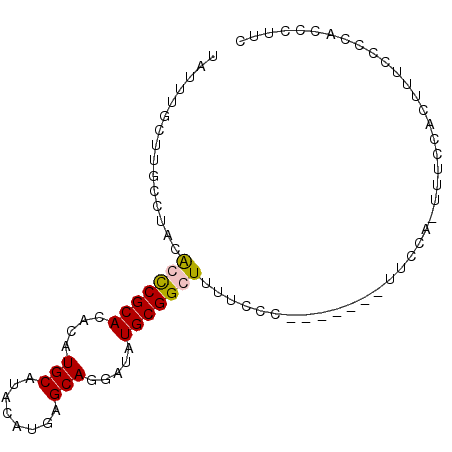
| Location | 2,019,054 – 2,019,145 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -11.18 |
| Energy contribution | -11.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2019054 91 - 22407834 UAUUUGCUUGCCUAGGCUCGCACACAUGCAUACAUGAGCAGGAUAUGCGGCUUUUCCCCUUCCAUUUUCA-UUUCCACUUCCCCCACCCUUC ....(((((((....))..(((....)))......)))))(((...(.((......))).))).......-..................... ( -12.80) >DroSec_CAF1 36580 84 - 1 AUUUUGCUUGCCUACAGCCGCACACAUGCAUACAUGAGCAGGAUAUGCGGCUUUUCCC-------UUUCA-UUUCCACUUUCCCCACCCUUC ...............(((((((....(((........))).....)))))))......-------.....-..................... ( -14.80) >DroSim_CAF1 35024 84 - 1 UAUUUGCUUGCCUACAGCCGCACACAUGCAUACAUGAGCAGGAUAUGCGGCUUUUCCC-------UUCCA-UUUCCACUUUCCCCACCCUUU ...............(((((((....(((........))).....)))))))......-------.....-..................... ( -14.80) >DroYak_CAF1 38230 73 - 1 UAUUUGCUUGCCCACGCUCGCACACAUGCAUACAUGAGCAGGAUAUGCGGCUUUCUCC-------UUCCACUUUCCACUU------------ ....(((((((....))..(((....)))......)))))(((...(.((........-------..)).)..)))....------------ ( -10.80) >consensus UAUUUGCUUGCCUACACCCGCACACAUGCAUACAUGAGCAGGAUAUGCGGCUUUUCCC_______UUCCA_UUUCCACUUUCCCCACCCUUC ...............(((((((....(((........))).....)))))))........................................ (-11.18 = -11.17 + -0.00)

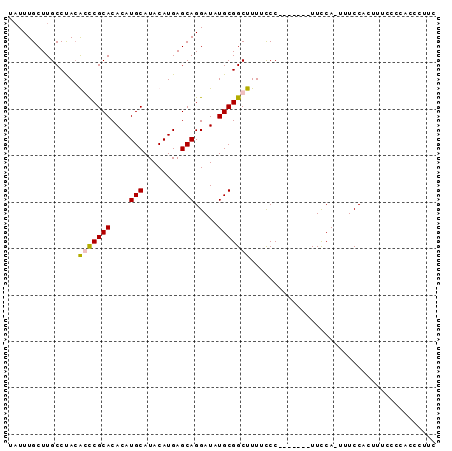
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:16 2006