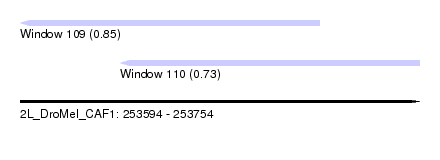
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 253,594 – 253,754 |
| Length | 160 |
| Max. P | 0.848744 |

| Location | 253,594 – 253,714 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -21.13 |
| Energy contribution | -20.22 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 253594 120 - 22407834 AACUACAUUAAGAAUGUGCGCCAGCACAUCAAGAUUCCAAUGCUUGCCAACGGAAACAUCCUGGCCCUGGACGACGUUCAUCGUUGUCUGACUGAAACAGGAGUCGAUGGUGUAAUGAGC ...(((((((.((.(((((....)))))))..((((((.......((((..(((....)))))))...((((((((.....))))))))..........))))))..)))))))...... ( -39.20) >DroPse_CAF1 64224 120 - 1 AGUUACAUACAGGAGGUGCGAAAGCACCUGAAAAUACCCAUGUUCGCCAAUGGCAAUAUACUGGGACUGGAGGAUGUCCACCGGUGUCUGGACGAGACGGGCGUGGAUGGUGUGAUGACU ((((((((((...((((((....))))))........(((((((((((...)))......(..(((((((.((....)).))))).))..).......))))))))...))))).))))) ( -49.30) >DroGri_CAF1 57970 120 - 1 AGUUAUAUUAAAGAUGUGCGCGAGCACAUUAAGAUACCCAUGCUUGCCAAUGGCAAUAUAUUGGGCGUGGAGGAUGUGCAUCGUUGUCUGACGGAGACGGGGGUCGAUGGUGUCAUGACU ((((((......(((((((....))))))).......((((((...((((((......)))))))))))).......(((((((((.((..((....))..)).))))))))).)))))) ( -46.50) >DroEre_CAF1 47385 120 - 1 AACUACAUCAAGAGUGUGCGGCAGCACAUCAAGAUUCCAAUGCUUGCCAACGGAAACAUCCUGGGCCUGGAGGACGUACACCGUUGCCUGGCGGAAACGGGAGUCGAUGGCGUAAUGAGC .((..((((..((.(((((....)))))))..((((((...(((.(.((((((..((.((((........)))).))...)))))).).)))(....).))))))))))..))....... ( -41.80) >DroYak_CAF1 48893 120 - 1 AACUACAUUAAGAGUGUGCGGCAGCACGUCAAGAUUCCAAUGCUGGCCAACGGAAACAUCCUGGGCCUGGAGGACGUACACCGUUGCCUGGCCGAAACGGGAGUCGAUGGCGUAAUGAGC .....(((((.....((((....))))((((.((((((.....(((((...(((....))).((((.(((.(......).)))..))))))))).....))))))..)))).)))))... ( -41.10) >DroPer_CAF1 65043 120 - 1 AGUUACAUACAGGAGGUGCGACAGCACCUGAAAAUACCCAUGUUCGCCAAUGGCAAUAUACUGGGACUGGAGGAUGUCCACCGGUGUCUGGACGAGACGGGCGUGGAUGGUGUGAUGACU ((((((((((...((((((....))))))........(((((((((((...)))......(..(((((((.((....)).))))).))..).......))))))))...))))).))))) ( -46.30) >consensus AACUACAUUAAGAAUGUGCGACAGCACAUCAAGAUACCAAUGCUUGCCAACGGAAACAUACUGGGCCUGGAGGACGUACACCGUUGUCUGGCCGAAACGGGAGUCGAUGGUGUAAUGACC .............((((((....)))))).................(((.(((.......)))....)))....((((((((((((((((.......))))...)))))))).))))... (-21.13 = -20.22 + -0.91)

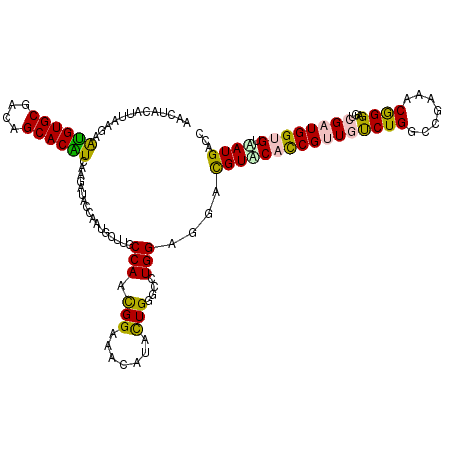
| Location | 253,634 – 253,754 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -27.91 |
| Energy contribution | -26.63 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 253634 120 - 22407834 CAGAGAGCAGAAGGGUCCGCUGACCGGGGUGGCUAAUUGGAACUACAUUAAGAAUGUGCGCCAGCACAUCAAGAUUCCAAUGCUUGCCAACGGAAACAUCCUGGCCCUGGACGACGUUCA ....((((......(((((..(.((((((((((..(((((((((......)).((((((....)))))).....)))))))....)))...(....)))))))))..)))))...)))). ( -40.40) >DroPse_CAF1 64264 120 - 1 CAGAGAGCAGAAGGGACCCCUGACAGGUGUGGCCGAUUGGAGUUACAUACAGGAGGUGCGAAAGCACCUGAAAAUACCCAUGUUCGCCAAUGGCAAUAUACUGGGACUGGAGGAUGUCCA (((.(((((...(((...((((....((((((((.....).))))))).))))((((((....)))))).......))).)))))(((...)))......)))((((........)))). ( -46.80) >DroGri_CAF1 58010 120 - 1 ACGUGAGCAGAAGGGUCCACUUACCGGCAUAGCCGAUUGGAGUUAUAUUAAAGAUGUGCGCGAGCACAUUAAGAUACCCAUGCUUGCCAAUGGCAAUAUAUUGGGCGUGGAGGAUGUGCA ..(((..(......)..)))......(((((.((..................(((((((....))))))).......((((((...((((((......)))))))))))).)).))))). ( -38.40) >DroEre_CAF1 47425 120 - 1 CAGAGAGCAGAAGGGUCCGCUGACCGGGGUGGCCAAUUGGAACUACAUCAAGAGUGUGCGGCAGCACAUCAAGAUUCCAAUGCUUGCCAACGGAAACAUCCUGGGCCUGGAGGACGUACA ..............((((.(((.((((((((((..((((((.....(((..((.(((((....)))))))..)))))))))....)))...(....))))))))...))).))))..... ( -38.80) >DroYak_CAF1 48933 120 - 1 GAGAGAGCAGAAGGGUCCACUGACCGGGGUGGCCGAUUGGAACUACAUUAAGAGUGUGCGGCAGCACGUCAAGAUUCCAAUGCUGGCCAACGGAAACAUCCUGGGCCUGGAGGACGUACA ..............((((.(((.(((((((((((((((((((((......)).((((((....))))).)....)))))))..)))))...(....))))))))...))).))))..... ( -40.20) >DroPer_CAF1 65083 120 - 1 CAGAGAGCAGAAGGGGCCCCUGACAGGUGUGGCCGAUUGGAGUUACAUACAGGAGGUGCGACAGCACCUGAAAAUACCCAUGUUCGCCAAUGGCAAUAUACUGGGACUGGAGGAUGUCCA (((.(((((...(((...((((....((((((((.....).))))))).))))((((((....)))))).......))).)))))(((...)))......)))((((........)))). ( -44.00) >consensus CAGAGAGCAGAAGGGUCCACUGACCGGGGUGGCCGAUUGGAACUACAUUAAGAAUGUGCGACAGCACAUCAAGAUACCAAUGCUUGCCAACGGAAACAUACUGGGCCUGGAGGACGUACA ..............((((.((..((((((((.(((.((((..((......)).((((((....)))))).................)))))))...))))))))....)).))))..... (-27.91 = -26.63 + -1.27)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:01 2006