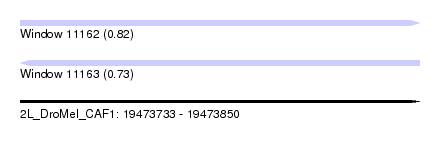
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,473,733 – 19,473,850 |
| Length | 117 |
| Max. P | 0.816347 |

| Location | 19,473,733 – 19,473,850 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -17.28 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
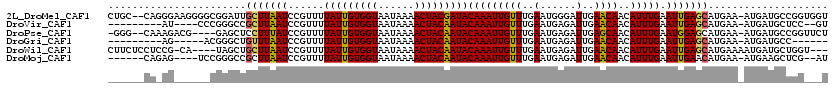
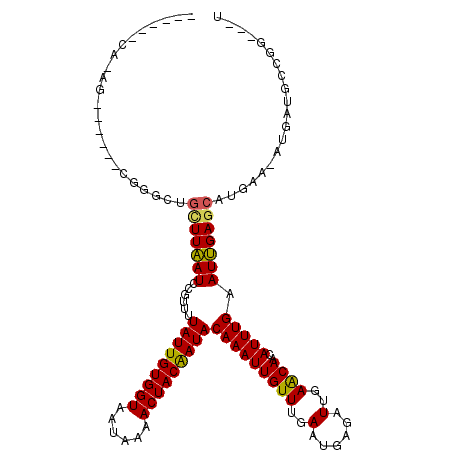
>2L_DroMel_CAF1 19473733 117 + 22407834 ACCACCGGCAUCAU-UUCAUGCUCAAUUCAAAUGUUGUUCAAUCCCAUUCAAACAAUUUGUAUCGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAAUCCGCCCCUUCCCUG--GCAG ......(((((...-...)))))..........((((((.(((((.......((.....))......((((((((.....))))))))))))).))))))..(((........)--)).. ( -20.70) >DroVir_CAF1 36169 104 + 1 AC--GGAGCAUCAU-UUCAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGG----AU--------- ..--.((((((...-...)))))).((((....((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))...))----))--------- ( -24.00) >DroPse_CAF1 36682 112 + 1 AGAACCGGCAUCAU-UUCAUGCUCCAUUCAAAUGUUGCUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUAAAGGAGCUC----CGUCUUUG--CCC- ......((((....-.....(((((.......((.....)).....((((..(((((....))))).((((((((.....))))))))))))...)))))..----......))--)).- ( -22.73) >DroGri_CAF1 32314 99 + 1 ------GGCAUCAU-UUCAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAACAGCCCGU-----CU--------- ------(((((...-...)))))..........((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))....-----..--------- ( -19.00) >DroWil_CAF1 44849 112 + 1 ---ACCAGCAUCAUUUUCAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCUA----UG-CGGAGGAGAAG ---.(((((((.......)))))....((..((((((((.(((((.......(((((....))))).((((((((.....))))))))))))).)))))).)----).-..))))..... ( -21.70) >DroMoj_CAF1 41310 107 + 1 AU--CGAGCUUCAU-UUCAUGUUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCGGCCCGGA----CUCUG------ ..--.((((.....-.....))))...((....((((((.(((((.......(((((....))))).((((((((.....))))))))))))).))))))...))----.....------ ( -18.40) >consensus A___CCAGCAUCAU_UUCAUGCUCAAUUCAAAUGUUGUUCAAUCUCAUUCAAACAAUUUGUAUUGUAGUUUUAUUACCACAAUAAAACGGAUUAAGCAGCCCG______CU_UG______ ......(((((.......)))))..........((((((.(((((.......(((((....))))).((((((((.....))))))))))))).)))))).................... (-17.28 = -16.65 + -0.63)

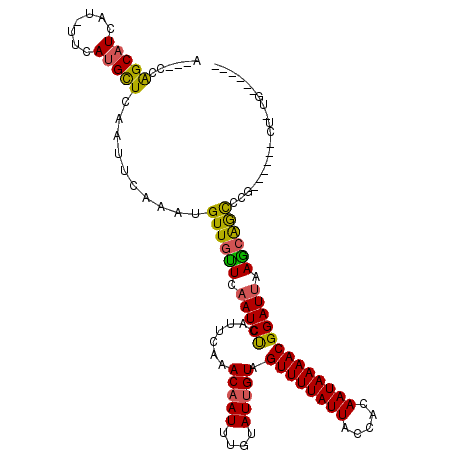
| Location | 19,473,733 – 19,473,850 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.53 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19473733 117 - 22407834 CUGC--CAGGGAAGGGGCGGAUUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACGAUACAAAUUGUUUGAAUGGGAUUGAACAACAUUUGAAUUGAGCAUGAA-AUGAUGCCGGUGGU (..(--(.......(((((((((....)))))))))(((((((((......)))))))))(((((((((..(......)..))))..)))))....(.((((...-...)))))))..). ( -29.90) >DroVir_CAF1 36169 104 - 1 ---------AU----CCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUGAA-AUGAUGCUCC--GU ---------..----..((((........))))...(((((((((......)))))))))(((((((((..(......)..))))..)))))....((((((...-...)))))).--.. ( -23.80) >DroPse_CAF1 36682 112 - 1 -GGG--CAAAGACG----GAGCUCCUUUAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAGCAACAUUUGAAUGGAGCAUGAA-AUGAUGCCGGUUCU -.((--((......----..(((((...........(((((((((......)))))))))(((((((((..(......)..))))..)))))...))))).....-....))))...... ( -28.03) >DroGri_CAF1 32314 99 - 1 ---------AG-----ACGGGCUGUUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUGAA-AUGAUGCC------ ---------..-----...((((((((((((((((((((((((((......)))))))))..........))))).))))))))).(((((.(........).))-)))..)))------ ( -23.90) >DroWil_CAF1 44849 112 - 1 CUUCUCCUCCG-CA----UAGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUGAAAAUGAUGCUGGU--- ........(((-((----(..(((((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).)))))))).)......)))).)).--- ( -25.90) >DroMoj_CAF1 41310 107 - 1 ------CAGAG----UCCGGGCCGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAACAUGAA-AUGAAGCUCG--AU ------.....----..(((((.(.((((((((((((((((((((......)))))))))..........))))).)))))).)..(((((.(........).))-)))..)))))--.. ( -23.10) >consensus ______CA_AG______CGGGCUGCUUAAUCCGUUUUAUUGUGGUAAUAAAACUACAAUACAAAUUGUUUGAAUGAGAUUGAACAACAUUUGAAUUGAGCAUGAA_AUGAUGCCGG___U .......................(((((((......(((((((((......)))))))))(((((((((..(......)..))))..))))).))))))).................... (-18.86 = -18.53 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:54 2006