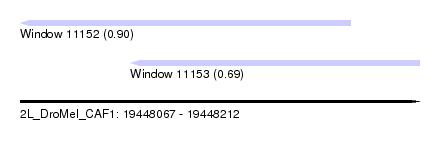
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,448,067 – 19,448,212 |
| Length | 145 |
| Max. P | 0.896601 |

| Location | 19,448,067 – 19,448,187 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -37.94 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.35 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19448067 120 - 22407834 AUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUUCACCAAGGAGCUGAUGUGGGUGUAUGACAACUCCCAAAAC (((.....(((((((.((..(..((((((((.(((.....))).)))))))).)..)).)))))))((.(((.(((......))).))).))))).((((.((......)).)))).... ( -45.70) >DroVir_CAF1 4840 120 - 1 AUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCAUAUUCUCUUGCGUCCGGAUUCGUAUAUUGGAUCCGUGGAGUUUACCAAGGAGCUAAUGUGGGUACAUGAUGUCGAAAAGAAU .((((.((.(((((((.........((...((((((((........))).)))))...))((((((((.(((.(((......))).))).)))))))))))))))..))))))....... ( -39.60) >DroGri_CAF1 4067 120 - 1 AUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCACAUUCUGUUGCGACCGGACUCGUAUAUUGGUUCCGUUGAGUUUACCAAGGAGCUGAUGUGGGUUCUUGAUCUUGAGAAAAAU .((((((((((((.((((..(....)....)))).)))...)))))).)))..((((((..((((..(((((.(((.(....)))))))))..))))))))))................. ( -35.60) >DroWil_CAF1 4342 120 - 1 AUCGAGCAGAUGUAUCAGAAGAAGACCCAAUUGGAGCAUAUUCUACUACGUCCCGAUUCGUAUAUUGGCUCCGUUGAGUAUACCAAAGAGCUAAUGUGGGUCUAUGAUUUCGAAAACAAU .(((((.......((((.....((((((((((((..(.((......)).)..))))))...((((((((((..(((.(....)))).)))))))))))))))).)))))))))....... ( -31.71) >DroMoj_CAF1 4076 120 - 1 AUCGAGCAGAUGUAUCAGAAAAAAUCGCAAUUGGAGCACAUCUUGUUGCGUCCAGAUUCGUACAUUGGCUCGGUGGAAUUUACAAAGGAGUUAAUGUGGGUACACGAUGCAGAAAAGAAU .((..(((..((((((.(((.....((((((.(((.....))).))))))......))).(((((((((((..((.......))...)))))))))))))))))...))).))....... ( -34.10) >DroAna_CAF1 3976 120 - 1 AUCGAACAGAUGUACCAGAAAAAGUCCCAGCUGGAACACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCUGUGGAGUACACCAAGGAGCUGAUGUGGGUGUAUGACUUUGAGAAGAAC .(((((...(((((((......(((((((((.(((.....))).)))).....)))))..(((((..(((((.(((......))).)))))..))))))))))))...)))))....... ( -40.90) >consensus AUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCCGUGGAGUUUACCAAGGAGCUAAUGUGGGUACAUGAUGUCGAAAAGAAU .((((....(((((((........(((((((.(((.....))).))))))).........((((((((((((...(......)...)))))))))))))))))))....))))....... (-25.82 = -26.35 + 0.53)


| Location | 19,448,107 – 19,448,212 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -21.17 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 19448107 105 - 22407834 GGAGAACGGAAACAAG---G------------CCCUGUCCAUCGAACAGAUGUACCAGAAGAAGUCGCAGCUGGAGCACAUCCUGCUGCGACCCGACUCGUACAUCGGAUCCGUGGAGUU ((.(...(....)...---.------------)))..(((((.((.(.(((((((.((..(..((((((((.(((.....))).)))))))).)..)).))))))).).)).)))))... ( -40.70) >DroVir_CAF1 4880 108 - 1 GGAGAAUGGUAAAGGUAAAG------------GCAUGUCCAUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCAUAUUCUCUUGCGUCCGGAUUCGUAUAUUGGAUCCGUGGAGUU ((((((((((..........------------.......((((.....))))..((((..(....)....)))).)).)))))))).((..(((((((((.....)))))))).)..)). ( -28.20) >DroGri_CAF1 4107 105 - 1 GCAGAACGGUAAAAAC---G------------CCAUGUCCAUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCACAUUCUGUUGCGACCGGACUCGUAUAUUGGUUCCGUUGAGUU ((.(((((((......---)------------))..((((.((((((((((((.((((..(....)....)))).)))...)))))).)))..))))..........)))).))...... ( -29.20) >DroWil_CAF1 4382 114 - 1 GGAAAACGGUAGCGGA---AAAUCUGGUG---GAAUGUCCAUCGAGCAGAUGUAUCAGAAGAAGACCCAAUUGGAGCAUAUUCUACUACGUCCCGAUUCGUAUAUUGGCUCCGUUGAGUA ....(((((.(((.((---......((((---(.....)))))((((.((((((..((((...(..((....))..)...))))..))))))..).))).....)).))))))))..... ( -29.90) >DroMoj_CAF1 4116 105 - 1 GGAGAACGGCAAAGGC---G------------CCAUGUCCAUCGAGCAGAUGUAUCAGAAAAAAUCGCAAUUGGAGCACAUCUUGUUGCGUCCAGAUUCGUACAUUGGCUCGGUGGAAUU .......(((......---)------------))...((((((((((.(((((((..(((.....((((((.(((.....))).))))))......)))))))))).))))))))))... ( -40.00) >DroAna_CAF1 4016 117 - 1 GGAGAACGGCAGCAAG---GCCUCUGGCGGGGCCAUGUCCAUCGAACAGAUGUACCAGAAAAAGUCCCAGCUGGAACACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCUGUGGAGUA ........((.....(---(((((....))))))...(((((.((.(.(((((((.((.....(((.((((.(((.....))).)))).)))....)).))))))).).)).))))))). ( -40.50) >consensus GGAGAACGGUAAAAAC___G____________CCAUGUCCAUCGAACAGAUGUACCAGAAGAAAUCGCAGCUGGAGCACAUCCUGCUGCGACCGGACUCGUACAUUGGCUCCGUGGAGUU .....................................(((((.((.(.(((((((.((......(((((((.(((.....))).))))))).....)).))))))).).)).)))))... (-21.17 = -20.70 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:45 2006